PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
Combine or Partition Data Matrices
The Combine or Partition Data Matrices... item in the Model menu is used to create a new data matrix from combining data matrices, or partitioning one or more existing data matrices.
By default, the initial screen will display the current data matrix (the last data matrix that was imported using the Import Spectroscopic Data Matrix... item in the Model menu).
There is an import button for each row that can be used to specify other CSV data matrices. Click on the first row where you wish to begin to add file sources. You can select multiple data matrix files and these will be appended to the dialog at the position associated with the button that was clicked to initiate the file import.
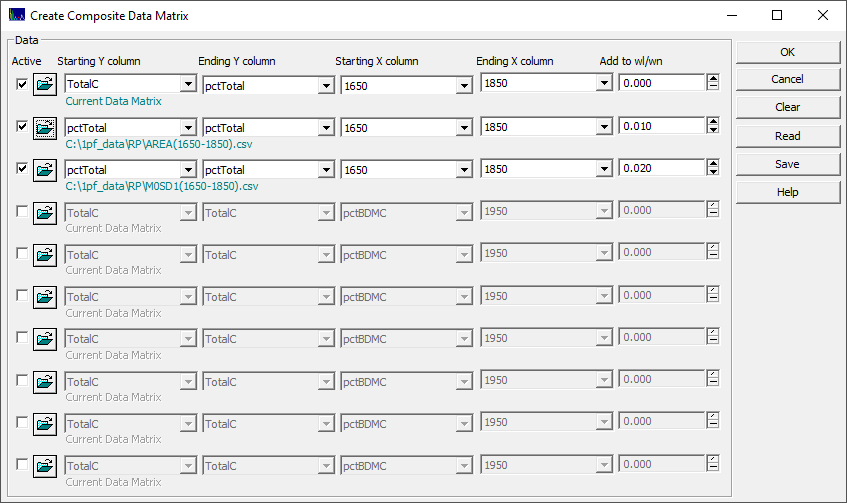
You must specify a starting and ending Y-column, and a starting and ending X-column for each entry. If you are importing multiple files with the same WLs, the optional Add to WL/Wn field can be used to differentiate which of the file sources is furnishing a given predictor. In the example below, two file source data matrices are specified. Each have unit wavelengths in the data matrix. To the first import, 0.01 is added to the wavelength, and to the second 0.02 is added.
You must be sure each of the line items to be used for the new data matrix are Active and properly specified.
When you are satisfied everything is specified, click OK, enter the name of the new CSV data matrix to be written. You will have an option to import this data and proceed to the fitting.

Combining Multiple CSV Data Matrices to Create a New CSV Data Matrix
You may have different normalizations, transforms, or principal components of your data that which you wish to combine into a single data matrix. For example, you may wish to fit a composite data set of the raw spectra, spectra that have been normalized to unit area, and spectra which have been normalized to a mean=0, standard deviation=1 in a single modeling. The example above does exactly this.
As another example, you may wish the fit the individual first through fourth principal component data reconstructions. In this case, you would import the first principal component data matrix (these can be easily generated in the Generate Principal Component GLM Data Matrices... option in the program) in the Import Spectroscopic Data Matrix... menu item, click on the import button in the second row of the specification, and import the three CSV files containing the additional principal components.
We have also included options to automatically generate center of mass and cumulative data matrices you may wish to combine with raw spectra.
You can specify the same data matrix multiple times, whether it is the current matrix or an imported one, for inserting multiple bands from each, but if imported, you must import the same file separately for each line in the specification where it is used.
When combining data sets, especially if you used the GLM Data option to save a file with outliers removed, you must be certain that all of the matrices from the different files have exactly the same count of spectra (rows), and that a given row in each of the files corresponds to the same identical sample. If you are managing the files separately in Excel, for example, be very cautious of the instance where certain files are sorted and others are not. The row counts must exactly match, and the samples represented in each row of the data matrix must match.
At least one Y-column is required for each line in the specification. Y-columns are used to check for row count and data integrity. You will need to repeat at least one of the Y columns, preferably the one that is most likely to be used for the fitting, in each line of the specifications. This will make it much easier to view the composite matrix in Excel and ensure that the samples are consistent across the different rows. In the example shown in the dialog, the pctTotal column will appear three different times. These should be an exact match for one another.
Since the X-columns of each matrix are appended to one another, the X-columns are unrestricted, but you will need to be cautious of the X-spacing in the subsequent fitting. An X-spacing of 10 may make sense for one of the data matrices, and a different X-spacing may make sense for another. Since the speed of the full permutation fitting will be dependent on the number of X-columns and the X-spacing, you will want to be consistent across the data sets. Also, use the X-column specification judiciously. If you know only certain wavelengths are of value with each of the component data matrices, specify the narrowest range possible. This will allow for the fastest possible fitting for a given X-column spacing.
The Generate Unit X-Spacing Data Matrix... option may be of value if the raw spectra have a high sampling density and you would prefer to have the data at unit wavelengths. You should do this before using any of the program's built-in options to modify the data matrix.
Although this option does require that a data matrix be currently loaded, the composite data set can consist only of imported CSV files. The current data matrix does not have to be used in any of the specifications.
Partitioning the Current Data Matrix to Create a New CSV Data Matrix
There may be instances where you want to include multiple specific WL bands from the current data matrix, and this option will allow up to ten included bands to be specified.
As with the combination of matrices, at least one Y-column is required for each line in the specification. For specifications beyond the first, simply repeat one of the Y-columns from the current matrix.
If you have more than ten bands, you can export a new composite matrix with the first ten specifications and import it with the Import Spectroscopic Data Matrix... item. By invoking the The Combine or Partition Data Matrices... item a second time with the new matrix, you can specify another ten bands. Alternatively, you can simply save the current data matrix file to a different name in Excel and delete the columns manually that you do not want in the predictors.
 |