PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
Subtract Baseline
This is PeakLab's baseline preprocessing option, used to remove the baseline prior to peak placement and fitting. This is accessed via the Subtract Baseline button in the main data window or the Subtract Baseline item in the main Data menu.
![]() You can also use the Subtract Baseline icon in the main program toolbar.
You can also use the Subtract Baseline icon in the main program toolbar.
In PeakLab, a baseline is identified, fitted, and then subtracted from the data. Since the baseline subtraction, especially with high S/N data, can impact the overall fit, there is no one-size-fits-all baseline subtraction. In general, there are just a few key points you will need to understand to remove a baseline to a high degree of accuracy.
This option offers baseline preprocessing that varies from close to wholly automated, to fully under a point by point manual control. This interactive baseline procedure also allows you to assign data peaks and data features as baseline and to remove them completely from the fitting. This can be useful if there are messy elutions within the middle of the data which are not of analytical interest and which could compromise the quality of the overall fitting.
Fitting a Baseline with the Peaks vs. Preprocessing
You can deal with a baseline in two ways. You can choose to fit the baseline along with the peaks, or you may elect to fit and subtract the baseline prior to peak fitting.
In modern chromatographic data sets, it is strongly recommended that the baseline be removed prior to fitting. This is a change from our recommendation when this technology first came to market almost three decades ago.
PeakLab uses new algorithms to allow very small magnitude peaks to be fitted alongside much more prominent ones. Fitting a baseline means that, on any given non-linear fitting iteration, a baseline can assume values which may totally override the presence of these smaller peaks. Further, as detectors and flow and temperature controllers have improved, baselines can often no longer be represented by simple models. In many cases, one may be looking at oscillating baselines arising from the small variations in these controllers. As small as these differences are, they can impact the overall goodness of fit when you are looking at near perfect fits with just a few ppm unaccounted variance, something unheard of those decades ago. Such near-perfect fits are now possible and were frequently realized in the development of PeakLab's fitting algorithms and models.
When fitting only a few peaks, each of a prominent area, and with a clearly defined linear baseline, it is probably safe to omit this preprocessing step and include the linear baseline in the overall model being fitted. You will need to use the Hidden Peaks - Residuals or Hidden Peaks - Second Derivative fitting options to fit a baseline alongside the peaks. The inclusion of a baseline is not offered in the simpler Local Maxima Peaks fitting option, which may be the only option you ever use if you never address hidden peaks that exhibit no local maxima.
When fitting a considerable number of peaks, especially when a broad range of areas are represented, we strongly recommend removing the baseline prior to fitting. This preprocessing step offers a non-parametric fitted baseline that can manage any baseline, however unusual its shape. Such shapes cannot be managed when the baseline is fitted alongside the peaks. Another advantage is that pre-subtracting the baseline simplifies the fit somewhat and in reducing the degrees of freedom, the fit statistics may be slightly improved.
The primary advantage of fitting baseline and peaks together was always that no additional error would be introduced by a separate baseline subtraction, but that assumed a simple model could be used to accurately describe the baseline and that fitting the baseline didn't interfere with the intricate fitting of low area components. When fitting many peaks, baselines will often shift away from an obvious baseline position in order to compensate for very small peaks near the baseline that are not being fitted as a consequence of the amplitude threshold.
Copying a Baseline State across Multiple Similar Data Sets
If you are processing multiple data sets where the baselines should be very close to identical across all or most of the data sets, it may be simplest to manually section the first data set or whichever one is most representative of the remaining sets. Once that baseline has been specified, you can use the right click menu in that specific graph for that data set and select the Copy this Baseline/Non-Baseline State to all Data Sets. This will produce the same baseline points specification for all of the data sets, although each will be individually fitted and subtracted. You can then zoom in each of the data set graphs to ensure each of the baselines are exactly where you would want them to be.
You may wish to use the Copy this Data Set's Baseline/Non-Baseline State to first put that baseline point state specification on the clipboard. You can then use the Paste Copied Baseline/Non-Baseline State to this Data Set right click menu option for each data set where this baseline points specification will be valid.
Algorithm
The Algorithm options apply specifically to the identification of the baseline points.
Initial,Final Linear
This is the simplest option. It simply selects the first and last active points, and constructs a line between them. The Numeric option is not available when this simple baseline is chosen since there are no fit statistics to report.
Progressive Lin
This option is appropriate for those data sets where it is known that a clear linear baseline exists at each end of the active data. PeakLab begins at the two limits and progressively fits a linear model to increasingly more points until the goodness of fit begins to deteriorate, usually signaling the first appearance of a peak. There are two progressive fits, one that works in an ascending direction, and one in a descending direction. The direction offering the better goodness of fit is automatically chosen. The Numeric option will display the fit statistics. For certain data sets, the ascending and descending statistics may be identical.
2nd Deriv Zero
This is the workhorse algorithm, the advanced baseline processing that has been in place since the first version of the product. The general principle is that baseline points tend to exist where the second derivative of the data is both constant and zero. For this 2nd Deriv Zero algorithm, you must select the count of points in the smoothing algorithm used to generate the second derivative and you must also specify a threshold for the baseline.
This D2 algorithm tends to be highly sensitive, more accurately detecting the start and end of peak, but there may be gaps in the assigned baseline points in zones that are pure baseline.
SD Variation
This baseline processing algorithm assumes that baseline points exist when a moving window SD across the data sees a increase beyond a specified threshold. Here you must also specify a point count for the smoothing. That same count is used for window of the SD computations. Here as well you must specify a threshold for the assignment of baseline points.
The SD algorithm tends to be less sensitive in capturing the precise start and end of a peak, but it will be very stable in assigning baseline points in zones that are pure baseline.
For sharply sloping spectral and gradient HPLC baselines where the start of the data and the end of the data have very different signal magnitudes, you will probably find that the 2nd Deriv Zero algorithm works best for automatic baseline specfication.
Parameters
The Parameters fields apply to the identification of baseline points when the the 2nd Deriv Zero or SD Variation algorithms are selected.
Sm n
For the 2nd Deriv Zero algorithm the Sm n specifies the count of points used to generate the smooth second derivative of the data using the Savitzky-Golay algorithm. The actual window count will be 2n+1, the center point plus n points on each side.
For the SD Variation algorithm, the Sm n specifies the count of points used to generate smoothed data using the Savitzky-Golay algorithm. Again, the actual window count will be 2n+1, the center point plus n points on each side. This Sm n is also the count of data points used in the sliding window computing the standard deviations. The SD window will be n points in length.
Adjust Threshold %
For both the 2nd Deriv Zero and the SD Variation algorithm, the Adjust Threshold % option specifies a percent of the maximum D2 magnitude or maximum SD as the baseline threshold. Typical values are around 1%. Generally, the greater the noise in the data, the greater will be the required tolerance. For gradient HPLC baselines, this value may be quite high.
The Adjust Threshold % option is useful when processing multiple data sets where the amount of baseline differs significantly across the different sets.
Adjust Baseline %
For both the 2nd Deriv Zero and the SD Variation algorithm, the Adjust Baseline % option specifies a percent of the points which shall be assigned to the baseline. The algorithm's threshold is determined in a linear optimization to get as close as possible to the specified value without exceeding the specified percentage. Note that specifying 50% for example, doesn't ensure that there will be exactly 5 of every 10 points assigned to the baseline. It should, however, be just shy of such if the Consec consecutive point requirement is set to 0. If Consec is set to some count, the percentage will likely be reduced.
This Adjust Baseline % option is useful when processing multiple data sets where there are significant S/N differences across the different sets, but the amount of peak and baseline are close to the same in each.
The actual % of baseline points is reported in the %BL = value shown the graph titles.
Model
The Model items apply only to the fitting of the points identified as baseline. Only the points identified as baseline are fitted. The fitted baseline is then subtracted from the incoming data.
When the 2nd Deriv Zero or SD Variation algorithms are selected, you must choose the model to be used for fitting the baseline:
· Constant (y=a0)
· Linear (y=a0+a1*x)
· Quadratic (y=a0+a1*x+a2*x2)
· Cubic (y=a0+a1*x+a2*x2+a3*x3)
· Logarithmic (y=a0+a1*ln(x))
· Exponential (y=a0+a1*exp(-x/a2))
· Power (Power, y=a0+a1*xa2)
· Hyperbolic (y=a0+a1*a2/(a2+x))
· Cnstrnd Cubic Spline
· Non-Parm Linear
The exponential, power, and hyperbolic baselines are non-linear models and are fitted iteratively using the same Levenburg-Marquardt algorithm used in peak fitting. The constant, linear, quadratic, cubic, and logarithmic baselines are linear and are fitted in a single step matrix solution. All fits use least-squares minimizations. These models fit the total of the identified baseline points; there is no local fitting.
The Cnstrnd Cubic Spline option creates a smoothed data stream using the Savitzky-Golay algorithm (the Sm n value is also used for this smoothing) and then creates a constrained cubic spline using the smoothed data. The constraint prevents the oscillations of the spline above and below the data points that can occur with unconstrained cubic interpolants. The smoothed data uses all of the data points, but the spline uses only the points identified as baseline. The spline algorithm will thus have to contend with the large gaps corresponding with peaks, treating the data as if it were equally spaced. In some cases, especially with very high S/N data, this option will produce a lovely trace of the non-linearity below the peaks when an oscillating baseline (a baseline with minima and maxima) is present. Because just four points enter into each estimate in the spline interpolant, the points on each side of a peak are critical to the shape that occurs below the peaks. When this procedure works, you will get a better estimate of the baseline below peaks, possibly a full cubic accuracy if the constraint isn't activated across the sampling gap in the baseline points.
The Non-Parm Linear option uses a PeakLab-specific non-parametric algorithm similar to Loess, but which in v5 now employs a uniform weighting function. This algorithm can fit any shape baseline if defined by sufficient points. This algorithm was used almost exclusively in our development work. It is stable, robust, and reasonably accurate insofar as the linear fit makes it very difficult to introduce anomalies, especially when a significant count of points is used as the NP n, the count of points used in the non-parametric algorithm's sliding window.
NP n
Wen the Non-Parm Linear algorithm is selected, you must specify the NP n, the count of points used in the sliding linear fitting window. A fit of n points occurs to get a single estimate of the baseline at a given x value. The non-parameteric procedure is thus computationally slower than the other types of fits. If you are simultaneously processing a dozen data sets in this baseline procedure, each with 10,000 data points, over 100,000 linear fits must be made each time you make any adjustment to the baseline identification or modeling. This NP n setting applies only to the Non-Parm Linear fitting. If your data consist of relatively few points, you can still use this algorithm, but you may need to set NP n to 3, the minimum.
Consec
As with any algorithm susceptible to noise, there can be benefits in requiring a count of consecutive points. The Consec option sets the count of points that must consecutively exist in order to be assigned as baseline. If Consec is set to 10, and the identification algorithm finds a string of nine consecutive points as baseline, with data on each side, the nine points are reassigned as valid peak data. The Consec option has no effect on points originally assigned as peak data; if even a single point is assigned to a peak, such is retained. This setting aplies to any of the models fitted using the baseline identified by the 2nd Deriv Zero or the SD Variation algorithms.
Post-Processing
Zero Baseline Points
The Zero Baseline Points will zero all data points identified as baseline instead of subtracting the model from the data. This is of significant value with the Cnstrnd Cubic Spline and Non-Parm Linear options when you manually assign unwanted peaks or other data features to baseline. It can be seen as the equivalent of erasing peaks of no interest from data. This is generally preferable to introducing unevenly spaced data in the Section Data option by sectioning away peaks or data features in the middle of the data sequence. For example, Fourier processing, such as the IRF Deconvolution, requiring uniformly sampled data, is not invalidated by this Zero Baseline Points option.
Zero Negative Points
The Zero Baseline Points will zero all data in the baseline subtraction that have negative values. Since this introduces bias in the fitting, it isn't typically done.
Graph - Scale Y
Visualizing the baseline is important. There are three scaling options for the Y-axis of the plots:
Baseline Points
This option scales for the points currently identified as baseline in each of the data sets. In most cases, this default scaling will show you the fine detail of the baseline being fitted.
All Points
You can use this option to scale for the whole of the data, baseline and peaks.
Common Baseline Points
This option is used when multiple data sets are being simultaneously processed. The scaling is based on the maximum and minimum Y values of the baselines across all of the sets. It can be useful for comparing baselines across different samples, both in absolute magnitude of the signal and in the specific shape of the signal drift.
Graph - Additional Y
Add D2 to Graphs
This option may be useful if you are using the 2nd Deriv Zero identification algorithm. This will plot the smoothed D2 data above the baseline, highlighting the smoothed second derivative points that qualify as baseline. The D2 algorithm is highly sensitive to insufficient smoothing levels. Too low a D2 smoothing results in the second derivative consisting of mostly noise. It is fairly easy to find the optimum D2 smoothing level when using this Add D2 to Graphs option. You may need to zoom-in the D2 data to see the necessary detail.
Manual Baseline Adjustment
![]() Be sure the Manual Sectioning Mode is selected in the graph's toolbar. This is the default when
the procedure is opened.
Be sure the Manual Sectioning Mode is selected in the graph's toolbar. This is the default when
the procedure is opened.
The automated procedures are often very effective at determining where a baseline exists, but they can be improved upon by human visual perception. You may see points that you know should be included in the baseline, and points that you know should not be included.
You may enable or disable a band of points using the same procedure as that used for Sectioning. To exclude active points (assign points as peak information), simply place the mouse cursor at the initial point to be excluded, click and hold the left mouse button down, and slide the mouse to the right. As you do so, the points will be grayed (indicating that they have been deactivated or excluded from the baseline fit). Once the left mouse button is released, the new fit will be made and the graph will be updated with the new baseline. To include a band of points (assign points as baseline information), simply start at the rightmost point desired, click and hold the left mouse button down, and slide the mouse to the left. As you do so, the points will be changed to an active state and used in the baseline determination.
In summary, baseline points are assigned by left clicking the mouse and sliding left and releasing the mouse when all of the intended baseline points have turned yellow (instead of gray). Peak points are assigned by left clicking the mouse and sliding right and releasing the mouse when all of the intended peak points have turned gray (instead of yellow).
You may need to use a manual baseline on messy elutions where the peaks do not decay to a clean baseline. To treat such 'drizzle' from the column as baseline, you may need to manually assign the baseline points.
Numeric
The Numeric option can be used to view the fit statistics for the baseline fit. The graph titles and the Numeric listing will display the r2 and F-statistic of the baseline fit as well as the r2, F-statistic, and ppm unaccounted variance limits arising from the noise or error in the baseline points in any subsequent fit to the baseline subtracted data.
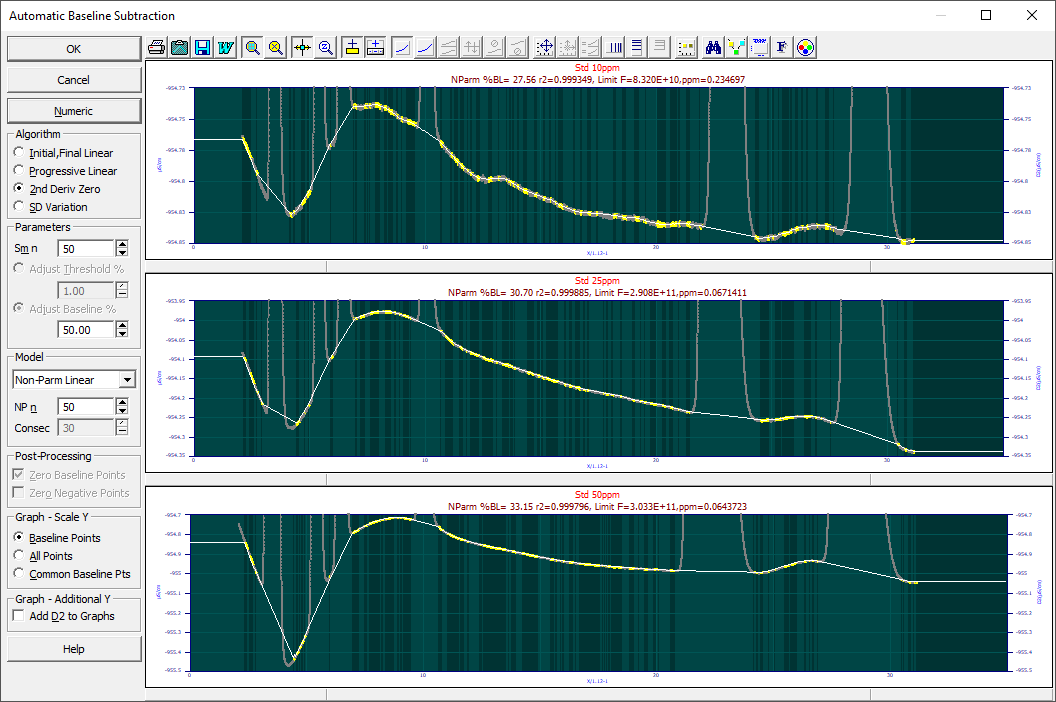
There will exist a certain fit error in any subsequent peak fit that arises from just the variance around the baseline of the points assigned as baseline. These best possible goodness of fit values achievable in any subsequent peak fit are the values you would see in these fit statistics if every point assigned to peak information had zero error in a subsequent fit. In the baseline processing dialog illustrated at the beginning of this topic, the noisier data in the uppermost graph, has a ppm limit of .23. This means the baseline points contain .23 ppm of unaccounted variance in any baseline corrected peak fit. If the final peak fit to the data has an error of 50 ppm unaccounted variance, it may help to know that the baseline fitting errors contributed an insignificant measure to the overall error.
Multiple Data Sets
If you have multiple data sets present, all will be shown in this procedure, irrespective of whether or not a data set is selected. The data set selection in the main window applies only to the View and Compare Data options and the Local Maxima Peaks, Hidden Peaks - Residuals, and the Hidden Peaks - Second Derivative fitting options.
When the OK is selected with all data sets shown, all of the data sets will be subtracted exactly as shown in the graphs and a new data level will be created containing the baseline corrected data.
To perform a custom baseline correction on one or more data sets, first explore the global dialog settings to get each baseline as close as you can. Then manually adjust each data set that requires fine tuning using the mouse within its graph. Note that the settings in the dialog are global. If you adjust any one of these settings, any custom baselines will be cleared.
If you need more detail, double click that graph or right click and select the Plot This Data Set option from the popup menu. Manually adjust that specific data set for the baseline points you wish and then click OK. The full set of baselines shown will now include this custom baseline. Note that only the overall baseline processing information is saved in the level description in the PFD file. A custom baseline notification will be given when you exit the procedure when one or more of the data sets were individually processed.
PeakLab Graph
All of the graph options are available in the PeakLab graph which is used throughout the program.
A Nod to Perfectionists
In the graphic first appearing in this topic to illustrate the dialog for this procedure, you will note the automatic baseline correction is less than perfect. While much depends on the magnitude of the baseline oscillations, and these are quite small, we will answer the obvious question with respect to this example data. How much better is a human version of the baseline correction done completely with the manual adjustment?
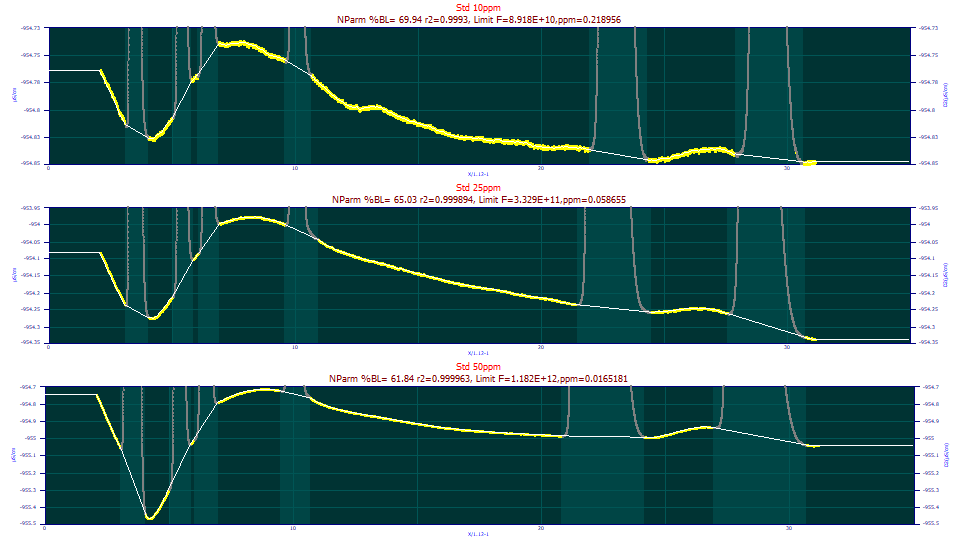
Visually, this manually adjusted non-parametric baseline looks near-perfect.
Here are the ppm unaccounted variance results from fitting the GenNLC<ge> model to the three sets using the automated and manual baseline corrections:
Baseline C=10ppm C=25ppm C=50ppm
Automatic 5.59 5.19 22.71
Manual 5.47 4.93 19.06
A 5 ppm unaccounted variance is an r2 of 0.999995. The F-statistic for the 10 and 25ppm concentrations of this standard were above 100 million (1E+8). This standard is real-world data containing six distinct peaks. Our takeaway is that the fits in PeakLab can be so close to complete in their accuracy that the subtleties in the baseline correction will impact the fit. On the other hand, that difference was in this case in the seventh decimal place of the r2 coefficient of determination of the fit. If your data is close to this same S/N (the noise floor in the Fourier Denoising procedure was -120, -140, -145 dB for these three data sets), and if you feel tempted to perform these near-ideal baseline corrections, you may be expending a considerable effort for very little benefit.
In developing the new models in PeakLab, we needed to have a reference for how far we were from a perfect statistical fit. The visualization in this Subtract Baseline option reflects such in this magnification of the nuances of baselines. If we are guilty of too much information, an exercise such as the one above should be sufficient for you to trust the automation. Once a baseline procedure has been established, the process only requires a few seconds.
Removing Unwanted Peaks or 'Messy' Features in the Data
This Subtract Baseline procedure is the method of choice for removing peaks or unwanted zones of information from data.
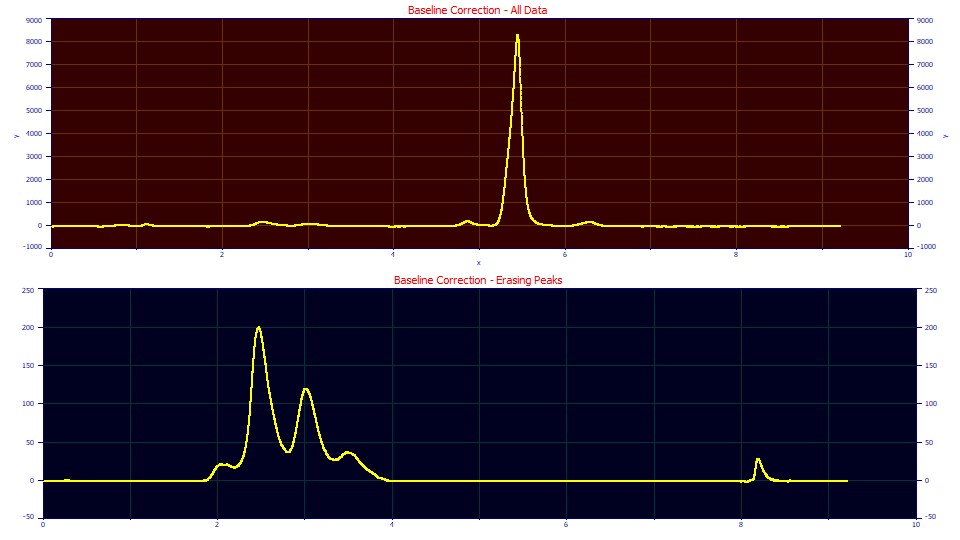
In the baseline procedure, you can 'erase' the peaks that have no interest in fitting. The upper plot consists of blood protein data where the principal peak dominates the chromatogram, but is of little analytical interest. The lower plot contains only the peaks of interest from this same data set. To remove unwanted peaks, you use the manual baseline adjustment. Place the mouse at the right side of such peaks to be erased, and then click and hold the left mouse button down while sliding the mouse left. This assigns the peak information in that sweep to the baseline. The Zero Baseline postprocessing option will zero all such information, 'erasing' it from the baseline corrected data.
A Starting Baseline Correction Strategy
(1) Start with the 2nd Deriv Zero baseline identification algorithm. You will probably need to use the SD Variation algorithm if you have HPLC gradient data. Where possible, we prefer the 2nd Deriv Zero for its greater threshold of sensitivity.
(2) For the D2 algorithm, set the Sm n to about .5% of the data set size. If the data consist of 10,000 samples, use a Sm n of 50. The D2 algorithm is highly sensitive to insufficient smoothing levels. Too low a D2 smoothing results in the second derivative consisting of mostly noise. Use the Add D2 to Graphs option if needed to find an optimal D2 smoothing level. For the SD Variation algorithm, the Sm n will typically be about one-half to one-third of this value.
(3) Choose the Adjust Baseline % and set it to 50% or whatever value visually appears to be approximately the percent of baseline points in the data.
(4) Use the Non-Parm Linear model and set its NP n to the same length as the smoothing Sm n. Set the Consec consecutive point requirement to about half this value. You will not likely want to use less than an NP n of 10, and you will probably want some value for Consec to prevent stray points in the decay of a peak from being treated as baseline.
(5) Adjust the Adjust Baseline %, and if needed, the Sm n smoothing, until you see an automated baseline correction that you are comfortable with.


 |