PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
Main Data Window
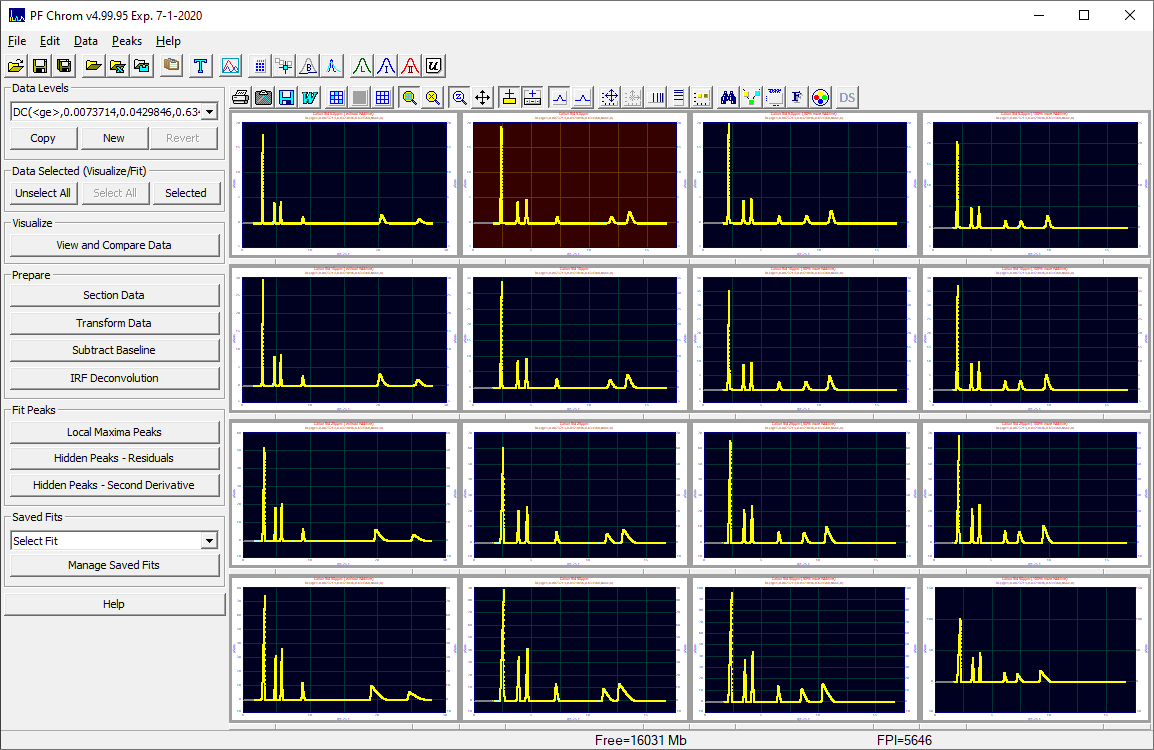
When data have been opened, the main data window presents most of the primary program functions:

Data Graphs
The data are shown graphically in an array of PeakLab graphs. There is a separate graph toolbar just above the graphs which can be used to control the different options in the graphs. PeakLab supports up to 25 data sets all of which can be simultaneously visualized, processed, and fitted. The data sets need not have the same X spacing or range, but we do not recommend mixing different x-scales such as minutes and seconds. Converting to k' retention units solves this issue.
Main Window Toolbar
The key import options and many of the primary program procedures are in the main toolbar.
Data Levels
A PeakLab data file can have as many as eight data levels. The first will always be the imported data or the data copied from an existing data level using the New option in order to create a new PeakLab data file. The subsequent data levels will be those arising from transforms, baseline correction, IRF deconvolution, and additional levels created using the Copy option on an existing level. In general there will be a progression of levels, such as imported data, a dead-time transform for retention time, a baseline correction, and an IRF deconvolution to remove the system distortions. The lowest data level will be the original. The additional levels are placed above, on top of, the prior levels. The current level in a sequence of data preparation will always be the topmost or uppermost level.
Uppermost Data Level Constraint
Most of the procedures in the program are available only to the data on the uppermost data level because they create a new data level in this sequential progression. Specifically, the Transform Data, Subtract Baseline, IRF Deconvolution options, as well as the specialty Smooth, Interpolate/Upsample, and Fourier Denoising options only available from the Data menu, each create a new data level and can only be invoked from this uppermost data level.
If you wish to create a separate transform, baseline, IRF, or other data level using data from an interior or non-uppermost level, you must first copy that existing interior level level to a new uppermost level using the Copy option. This maintains the coherence of the data preparation sequencing and prevents what could be a very large number of saved fits being invalidated in a single step.
The exceptions to this uppermost constraint are the Visualize and Compare Data option and the Local Maxima Peaks, Hidden Peaks-Residuals, and Hidden Peaks-Second Derivative fitting options. These can be used on any level since they do not change the data; they only visualize or fit it. Fits are assigned to the data level used for the fitting.
The Section option, which partitions data into active and inactive regions, is managed somewhat differently. Since it does not change the data values, only the states of the points in the data, this option applies individually to each data level and can be used on any level at any time. When a new data level is generated, it inherits the sectioning of the prior level. You can go back after the fact and alter the sectioning on any level, but this only changes that level. This will invalidate all fits assigned to that level since a fit will be specific to the active data (that which is used in the fitting). If you wish to create a second sectioned level of data for different fitting, you must use the Copy option and resection this new uppermost level in order to preserve existing fits from the prior sectioning.
Data Levels Dropdown
Use the dropdown to select the active data level. The data level name can be changed in the Titles option.
Copy
Use the Copy button to copy the currently selected data level to a new uppermost level. Any level can be used for the Copy. The new uppermost level can then be sectioned, given a different transform, baseline correction, IRF deconvolution, or other data processing.
New
Use the New button to copy the currently selected data level to a new PeakLab file where this data level is the only data in the file. This is useful if you find the eight levels insufficient for an analysis, or if you wish to create a data set containing only one specific level from the other file. You will be given an option to Save the current file before the new one is created. The new one becomes the current opened file.
Revert
Use the Revert button to erase all data levels subsequent to the currently selected one. This makes the current level the new uppermost level. Use this option cautiously since all fits on the deleted levels will also be erased. You will be given an option to save the current file before the erasure of the subsequent levels is done. Please do so if you invested a good measure of time in the fits that will be deleted and there is a chance you may wish to revisit them. As an alternative, you can use the File | Save option to save the current file, and the File | Save As to save the same file with a different name. You can then Revert the new file as you like.
Selecting Data Sets
Only selected data sets are used for visualization and fitting. In order to visualize or fit only certain data sets and not others, it is a simple matter to select and deselect data sets.
Using the mouse, simply click a graph to select or unselect it. A thick gray border indicates selection, a thin gray border indicates an unselected state. This will work on any data level. For the non-uppermost or interior data levels, the background color will be indicative of an interior level. An interior data level is deemed to consist of committed read-only data. By default, this will be a copper background color.
For the uppermost data level, the background will also change colors with selection and deselection. By default the background will be gray for unselected data sets, and blue for selected sets. There is one other data state on this uppermost data level and it will contain a red background. This is the 'principal' data.
Principal Data
The red-background principal data selection is useful for a number of program options which offer a specific export or edit for the principal data set. Note that the 'principal' data is treated no differently than any other data set. It is mainly important in user-defined peaks which use data set statistic functions such as YMAX in estimate formulas. These values will be based only on the principal data set. For most other matters, the principal data selection is mostly a convenience. Also, since every data file requires a principal data set, it is impossible to deselect all data sets.
Unselect All
Use the Unselect All button to deselect all data sets in single click. This will not deselect the principal data set. You may also right click any graph and choose Unselect All Data Sets from the popup menu.
Select All
Use the Select All button to select all data sets in single click. This will not affect the principal data set which is always selected. You may also right click any graph and choose Select All Data Sets from the popup menu.
Unselect or Select an Individual Data Set
Left click on the graph to select and deselect. You must do so without a motion that boxes a region of the graph. If you accidentally zoom-in all of the plots when clicking graphs on and off, use the Restore Scaling - Undo Zoom from the right click popup menu of any one of the graphs. To select and unselect an individual data set, you may also right click the specific graph and choose Unselect this Data Set or Select this Data Set from the popup menu.
Assigning the Principal Data Set
Right click the specific graph and choose Select as Principal Data Set from the popup menu. Note that the principal data set cannot be deselected on any data level and can only be changed on the uppermost data level.
Selected Popup
The Selected button opens a popup containing all currently selected data sets. The principal data will be highlighted in red. Click the Selected button again to close this popup or you can close it directly.
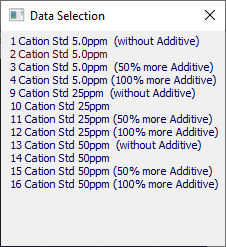
The same right click menu options available to the graphs are available from each of the entries in this popup.
To see the graph of just one of these data sets, click the mouse on that entry. Click again on the revised entry to restore all of the graphs.
You can reorder the data set sequence in the graphs, by clicking and holding the mouse on one data set and dragging it to the new position in the list
Color Scheme for Selected States
Right click a graph with the state you wish to change and select Choose Color Scheme for Unselected Data Sets..., or Choose Color Scheme for Selected Data Sets..., or Choose Color Scheme for Principal Data Set..., or Choose Color Scheme for Committed Read-Only Data Sets..., or Choose Color Scheme for Plot of Selected Data... from the popup menu.
Viewing One Data Set
When there are multiple data sets, you can double click one graph to view a only that graph full size. You can then double click that single graph to restore all of the plots.
You can also choose Plot | Only this Data Set (Disables Selection) from the right click popup menu to see this individual graph or Plot All Data Sets (Enables Selection) to restore all graphs.
You can also use the Selected popup as mentioned above.
Viewing Selected Data Sets
To see only the currently selected data sets, You can choose Plot | Only Selected Data Sets (Disables Selection) from the right click popup menu or Plot All Data Sets (Enables Selection) to restore all graphs.
Zooming-In Graphs
![]() In the graph toolbar, if the Zoom-In Applies to All Graphs is selected, the XY zoom in one graph
applies all of the graphs.
In the graph toolbar, if the Zoom-In Applies to All Graphs is selected, the XY zoom in one graph
applies all of the graphs.
![]() In the graph toolbar, if the the Zoom-In Applies to All Graphs, X-scaling only, the X zoom in one
graph applies all of the graphs. The Y-zoom is not used.
In the graph toolbar, if the the Zoom-In Applies to All Graphs, X-scaling only, the X zoom in one
graph applies all of the graphs. The Y-zoom is not used.
If neither button is depressed (selected), the zoom-in will apply only to the XY of the graph where you boxed the XY region.
The zoom is done by left clicking and holding the button, dragging the mouse to the opposite corner, and releasing the button.
To restore the default scaling, use the Restore Scaling - Undo Zoom from the right click popup menu of any one of the graphs. This will apply to all of the graphs.
XY Hints
![]() In the graph toolbar, the Show Graph hints will show the XY under the cursor. If this button is
selected for a second time, the hints will be synchronized in X across all of the graphs, and you will
see the location and Y values in each of the graphs. This is useful for seeing what is present at a given
retention value across multiple data sets. Clicking this button a third time will disable the hints.
In the graph toolbar, the Show Graph hints will show the XY under the cursor. If this button is
selected for a second time, the hints will be synchronized in X across all of the graphs, and you will
see the location and Y values in each of the graphs. This is useful for seeing what is present at a given
retention value across multiple data sets. Clicking this button a third time will disable the hints.
Reordering Data Set Sequence
![]() The Change Position Mode button in the graph toolbar is a mode where you drag each graph to the
location you wish in the graph matrix. You thus change the data sequence in the file. Simply click and
hold the left mouse button down and drag the peak to the location desired for that data set. This can
be done at any time, irrespective of the count of data levels and however many fits you may have already
done.
The Change Position Mode button in the graph toolbar is a mode where you drag each graph to the
location you wish in the graph matrix. You thus change the data sequence in the file. Simply click and
hold the left mouse button down and drag the peak to the location desired for that data set. This can
be done at any time, irrespective of the count of data levels and however many fits you may have already
done.
You can also use the Selected popup to reorder the data set sequence.
Graph Array Layout
To control the number of graphs displayed per row of the matrix, right click any graph and choose Specify Array Layout for Multiple Graphs... from the popup menu. For each count of total graphs 1-25, you specify the row count you wish to see. Certain of these selections may be overridden depending your video resolution and window sizes.
Removing One or More Data Set(s)
When your remove a data set, you remove the data from the PeakLab file. You do not delete the raw data that was imported into the program. When you right click a graph you will see two options Remove | This Data Set and Remove | All Unselected Data Sets. This cannot be undone; you may wish to save the existing data when offered.
If you wish to create a data file containing only a single data set, you may wish to make this the principal data set, use the Unselect All button, and then choose this Remove | All Unselected Data Sets option.
If you wish to create a data file containing only a subset of the current data sets, unselect each of those you wish to remove, and then choose this Remove | All Unselected Data Sets option.
All of the fits currently in the file associated with the retained data are preserved.
Data Titles
To change the titles for the individual data sets, for the data levels, or for the overall experiment use can use the right click Edit Data Titles... popup menu option.
![]() You can also use the Titles icon in the main program toolbar.
You can also use the Titles icon in the main program toolbar.
Each data level has its own data titles and its own level title. You must thus first select the data level of interest to make it the current one before selecting this titles option. Depending on the operation, titles may be inherited on subsequent data levels. You can go back and alter the titles on any level, but these are not propagated across the different levels that saw this inheritance. You will need to adjust the titles of each of the levels you wish to revise.
Note that PeakLab graphs have separate 2D custom titles and 3D custom titles options which may use some some portion of these data titles.
Edit Data
To manually edit the numeric data in the uppermost data level for any one of the individual data sets, select that data's graph and use the right click Edit this Data Set... | PeakLab Editor... or Edit this Data Set | ASCII Editor... popup menu option. The first option uses the PeakLab specific data editor and the second the ASCII text editor. When these editors are used the revised data replaces the original. The original is not preserved.
Data Export and Copy
Export this Data Set
To export a specific data set, right click the graph for the data set of interest and choose Export this Data Set... from the popup menu. You can save the data to disk as a [csv] comma-delimited ASCII file, as a [prn] tab delimited ASCII file, as a [txt] space delimited ASCII file, or as [cdf] AIA format net-cdf file.
The main File menu's Export Data Set(s) option offers three exports: Export Principal Data Set to ASCII CSV File..., Export Selected Data Sets to ASCII CSV File.., and Export All Data Sets to ASCII CSV File... for writing a CSV file that can be directly imported into Excel.
Copy this Data Set
To copy a specific data set to the clipboard, right click the graph for the data set of interest and choose Copy this Data Set... from the popup menu.
![]() The main Edit menu's Copy option offers three choices: Copy Principal Data Set, Copy
Selected Data Sets, and Copy All Data Sets for placing this tab-delimited information on the
clipboard.
The main Edit menu's Copy option offers three choices: Copy Principal Data Set, Copy
Selected Data Sets, and Copy All Data Sets for placing this tab-delimited information on the
clipboard.
Data Property Computations and Export
Export Data Property Computations...
To export specific properties of the data on the current data level to a CSV file, use the Export Data Property Computations... right click menu option. This is useful for generating predictors from spectral data you may wish to model to your chromatographic parameters.
Data Averaging, Subtraction, Replication
Averaging Two Data Sets
Use the Combine this Data Set with Principal Data to Export a Composite Data Set right click menu option to export a data set consisting of an average of this data set and the principal data set. You can save the composite data to disk as a [csv] comma-delimited ASCII file, as a [prn] tab delimited ASCII file, as a [txt] space delimited ASCII file, or as [cdf] AIA format net-cdf file.
Averaging Multiple Data sets
Use the Combine All Selected Data Sets to Export a Composite Data Set right click menu option to export a data set consisting of an average of all selected data sets. You can save the composite data to disk as a [csv] comma-delimited ASCII file, as a [prn] tab delimited ASCII file, as a [txt] space delimited ASCII file, or as [cdf] AIA format net-cdf file.
Subtract One Data Set from Another Data Set
If two data sets have identical active x-values, you can subtract one data set from another. This can only be done on a single initial data level. You can first section this data, but if you do this subtraction post-sectioning, you should use the Apply option in the Section procedure to ensure that each data set has the same exact count and selections of active x points. For this subtraction, you will need to assign the data set to be subtracted to the principal data. The Subtract Principal Data from this Data Set right click menu option will subtract this principal data set from the data set in the graph you have chosen. In this instance, this principal data file would normally be a blank. The revised data replaces this initial data level's data; the original data is lost. It is assumed the blank-subtracted data should be the input data.
Subtract One Data Set from All Other Data Sets
If data sets have identical active x-values, you can subtract one data set from all of the other data sets. This can only be done on a single initial data level. You can first section this data, but if you do this subtraction post-sectioning, you should use the Apply option in the Section procedure to ensure that each data set has the same exact count and selections of active x points. The Subtract this Data Set from All Data Sets right click menu option will subtract the data in the graph you have chosen from all other data sets in the file. The graph selected would normally be for a blank. The revised data replaces this initial data level's data; the original data is lost. It is assumed the blank-subtracted data should be the input data.
Replicate this Data Set a Specified Number of Times
This is usually used for Model Experiments with a single data set. The selected data set is replicated a specific number of times and these replicates are placed within the current data file. If you create 11 replicates of a single data set, you will have 12 identical data sets which can be fit to 12 different models simultaneously using the Model Experiment feature in the Local Maxima peak fitting.
Informational Options
Data Set Statistics
The right click Stats for this Data Set... menu option will display data table statistics. These will exist for each data set in each data level.
AIA netCDF File Information
Use the right click AIA File Information for this Data Set... popup menu option to view the AIA source information if the data originated from an AIA net CDF data file.
Assign Experimental Variables to Data Sets
To review experiments in the Explore option of the Review, you can assign experimental variables to each data set. Use the Map Experimental Process Variables for All Data Sets... right click menu option to assign experimental variables to each data set. This option is also available as a right click menu option in the Review. These variables are saved in the data file.
Add Location Reference Lines to Plots
The right click Specify X Location Line References for Graphs option is used for placing vertical reference lines and on a plot to identify specific expected elution times or spectral frequencies.
View and Compare Data
To visualize imported data or the processed data on any data level, select that level and highlight the data sets you wish to consolidate for visualization and then click the View and Compare Data button. You can also use the Data | View/Compare Data main menu option.
![]() You can also use the View/Compare Data icon in the main program toolbar.
You can also use the View/Compare Data icon in the main program toolbar.
This offers a host of 2D and 3D visualizations of the raw data. This option does not change the data table and is available to any data level. Only selected data sets are passed to the procedure. To view and compare different sets of data, simply select the ones of interest. These selected states also apply to fitting, so you will need to adjust your selections prior to fitting if those used for visualization do not represent those you wish to fit.
Section Data
The Section Data button or the (Data | Section menu item) opens the sectioning procedure. This is where the data is partitioned to an active region for fitting.
![]() You can also use the Section button in the main program toolbar.
You can also use the Section button in the main program toolbar.
All of the data sets are passed to the sectioning procedure irrespective of their selected state in the main window.. To section just a single data set, right click that data's graph and choose Independently Section this Data Set from the popup menu.
You can resection one or more data sets on a given data level after additional levels have been created, but you will lose any fits associated with this data level, and the changes will not propagate to these subsequent levels. The sectioning of each data level is independent, although newly created data levels will inherit the current state of the sectioning for each of the data sets.
Transform Data
The Transform Data option is a transform preprocessing procedure used to alter the data using user-entered calculations. This option is only available from the topmost data level. This procedure is accessed via the Transform Data button in the main data window or the Data|Transform Data in the main menu.
![]() You can also use the Transform Data icon in the main program toolbar.
You can also use the Transform Data icon in the main program toolbar.
All of the data sets are passed to the transform procedure irrespective of their selected state in the main window.. For transforming just one data set when many are present, right click the data set of interest and select Apply Transform to this Data Set from the popup menu.
The most common transform you are likely to use is the dead-time conversion to k' retention units. The program's full parser is available enabling you to create any computation you may need.
You cannot reaccess a transformed data level for additional transforms, or access it for any other procedure that further alters the data, once it is no longer the uppermost data level. Once a transform level is created, you should consider it a read-only data level. You can, however, Revert to a data level before this transform was done and initiate a new transform. If you do so, all fits in the discarded levels will be lost unless you save the data prior to the reversion.
Subtract Baseline
The Baseline option is a baseline preprocessing procedure, used to fit and remove the baseline prior to peak placement and fitting. This is accessed via the Subtract Baseline button in the main data window or the Data|Subtract Baseline in the main menu.
![]() You can also use the Subtract Baseline button in the main program toolbar.
You can also use the Subtract Baseline button in the main program toolbar.
All of the data sets are passed to the baseline procedure irrespective of their selected state in the main window.
Individual baseline corrections are done from within this Subtract Baseline procedure.
The procedure is a sophisticated fitting procedure that has been designed to fit a baseline to high accuracy for the best possible goodness of fit in subsequent peak fitting.
You cannot reaccess a baseline-corrected data level for revisions, or access it all once it is no longer the topmost data level, for any other procedure that alters the data. Once a baseline-corrected level is created, it is also a read-only data level. If needed, you can use the Revert option to undo one or more data levels to one before this baseline correction was done in order to redo the baseline. All fits in the discarded levels will be lost. If you fit a baseline level and you wish to explore a different baseline fit, you should Copy the data level prior to the baseline and use that for a second baseline correction level that you also fit. You can then access the saved fits from each level to compare the differences arising from the baseline preprocessing.
IRF Deconvolution
Fourier deconvolution is the procedure by which the IRF (instrumental distortions) are removed in a Fourier domain preprocessing step. The IRF Deconvolution button or the (Data | Deconvolve Instrument Response Function main menu item) opens the procedure for this IRF deconvolution preprocessing of data.
![]() You can also use the Deconvolve Instrument Response Function button in the main program toolbar.
You can also use the Deconvolve Instrument Response Function button in the main program toolbar.
Here, as well, all of the data sets are passed to the IRF deconvolution procedure irrespective of their selected state in the main window.
Individual IRF deconvolutions are done from within this IRF Deconvolution procedure.
The procedure is a highly sophisticated Fourier domain deconvolution and noise filtering procedure which allows you to import fitted values from convolution integral fits where the IRF parameters have been estimated. There is also a genetic algorithm that can often find a good IRF estimate for baseline resolved peaks. For many chromatographic peaks, it is necessary to either fit an IRF-bearing convolution integral model, or subtract a known IRF prior to fitting in order to obtain the highest accuracy fits.
Local Maxima Peaks
This peak fitting option is used primarily to fit local maxima peaks. Here peaks are identified strictly from their local maxima in a smoothed data stream. Local maxima peaks are those which have a visible apex, not necessarily those which are fully baseline resolved. The hidden peak procedures identify where those apex points are deemed to exist. If there is a clearly defined apex for each of the peaks, this Local Maxima Peaks fitting option is simpler. Unless you wish to search for hidden peaks prior to fitting, there in no need to use the more complex hidden peak fitting procedures. In most cases, an unaccounted hidden peak's presence will be apparent within a final fit, as in most peaks fitting quite well, and one or two with a clear anomaly in shape or discrepancy in parametric value trends.
This option is invoked from the Local Maxima Peaks button and from the Peaks | Fit Local Maxima Peaks main menu item.
![]() You can also use the Fit Local Maxima Peaks button in the main program toolbar.
You can also use the Fit Local Maxima Peaks button in the main program toolbar.
This is the only fitting option that offers the Model Experiment feature, making it easy to test replicates of a single data set against a variety of user-selected model candidates.
This option does not fit a baseline or allow the option of selecting a specific smoothing algorithm used for peak identification and initial estimates. For either of these you will need to use the Hidden Peaks - Residuals option. For most chromatographic data, we recommend subtracting the baseline prior to fitting.
Note that this Local Maxima fitting option can still fit hidden peaks with manual placement of such peaks, although in general, that placement will be aided by using one of the hidden peak fitting options.
To fit just a single data set, right click the graph for that specific data and choose Independently Fit this Data Set | Local Maxima... from the popup menu.
Hidden Peaks - Residuals
This peak fitting option is used primarily to fit hidden peaks.
This option is invoked from the Hidden Peaks - Residuals button and from the Peaks | Fit Hidden Peaks - Residuals main menu item.
![]() You can also use the Hidden Peaks - Residuals button in the main program toolbar.
You can also use the Hidden Peaks - Residuals button in the main program toolbar.
You can use this option, or the Fit Hidden Peaks Second Derivative option, to fit a baseline rather than removing such in a preprocessing step.
This is the only the fitting option where you can select the specific smoothing algorithm used for peak identification and initial estimates. This may be an issue with unusually noisy data where the default Savitzky-Golay smoothing algorithm produces artifacts in the data stream which are detected as peaks.
This Residuals option first identifies peaks by identifying local maxima in a smoothed data stream. This first step finds only those peaks which are visible peaks. PeakLab defines a visible peak as one which produces a local maximum in the data stream. A hidden peak is thus defined as one which fails to produce this local maximum. Note that peaks which lack this local maximum may range from being quite apparent in visual inspection to those which are altogether indiscernible. The hidden peaks are then optionally placed from the difference between the data and the estimated local maxima peaks (residuals).
Adding the peaks arising from the residuals is optional. This method can be used as a local maxima fitting procedure with the additional visualization of the residuals that may reveal hidden peaks. All of the capabilities within the Fit Local Maxima Peaks option are available in this method except the Model Experiment option.
To fit just a single data set, right click the graph for that specific data and choose Independently Fit this Data Set | Hidden Peaks - Residuals... from the popup menu.
Hidden Peaks - Second Derivative
This peak fitting option is also used to identity and fit hidden peaks.
This option is involved from the Hidden Peaks - Second Derivative button and from the Peaks | Fit Hidden Peaks - Second Derivative main menu item.
![]() You can also use the Hidden Peaks - Second Derivative button in the main program toolbar.
You can also use the Hidden Peaks - Second Derivative button in the main program toolbar.
You can use this option, or the Fit Hidden Peaks Residuals option, to fit a baseline rather than removing such in a preprocessing step.
In this option, hidden peaks are detected by second derivative minima. A peak that has no local maximum in the primary data, may very well have a local minima in a smoothed second derivative.
PeakLab defines a visible peak as one which produces a local maximum in the input data. A hidden peak is thus defined as one which fails to produce this local maximum. Peaks which lack this local maximum may range from being quite apparent in visual inspection to those which are altogether indiscernible.
This option uses the Savitzky-Golay algorithm to produce a smooth second derivative. As such, a constant x data spacing is required. If data lack this uniform x-spacing, or if artifacts or outliers have been excluded, this option can still be used, provided the data is first processed with PeakLab's Interpolate/Upsample option.
To fit just a single data set, right click the graph for that specific data and choose Independently Fit this Data Set | Hidden Peaks - Second Derivative... from the popup menu.
Storing Fits in a PeakLab Data File
You can fit the data on any data level at any time. Fitting the data does not alter the data in that level. In order to fit the data for a non-topmost data level, go to uppermost data level and select the data sets you wish to fit. Then select that level and initiate the fitting. Each fit of a given level's data is associated with that level, and counts as a single 'fit' in the data file, even if it consists of a fit for each of the data sets.
A PeakLab data file can contain 100 named fits, each containing the individual fits of up to 25 data sets. There can thus be as many as 2500 discrete fits in a PeakLab data file, and those can be associated with any number of data levels, or only a single one if that is where all of the fitting is done. You are offered the option of saving a fit at the conclusion of the Review process, when you know for certain you wish to save it. You must thus click the the Review Fit button at the conclusion of a fit in order for it to be saved.
Select Saved Fit
All fits assigned to the currently selected data level are listed in the Saved Fits dropdown. If you select a saved fit, you will be taken to the Review of that fit as if that fit had just been made. In fact, the fit is recomputed from the saved parameters as a data integrity measure. To see the fits assigned to a different data level, simply select that level in the Data Levels dropdown at the top of the dialog.
Manage Saved Fits
The Manage Saved Fits option opens a management dialog for the fits saved in the PeakLab file. It is used to rename and delete saved fits.


 |