PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
Chromatography Functions
HVL (Haarhoff-Van der Linde)
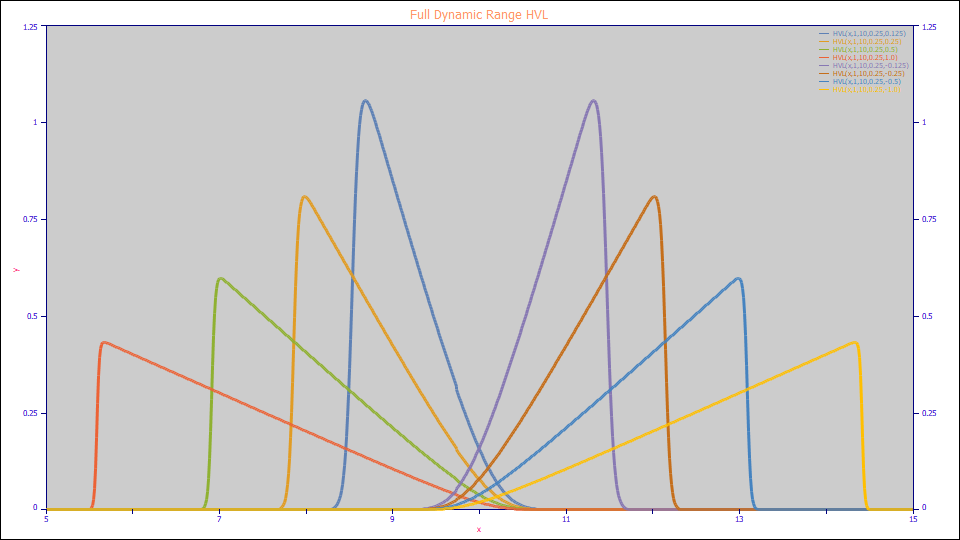
The HVL (full range) model:
a0 = Area
a1 = Center (as mean of asymmetric peak)
a2 = Width (SD of underlying Gaussian)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
Built in model: HVL
User-defined peaks and view functions: HVL(x,a0,a1,a2,a3)
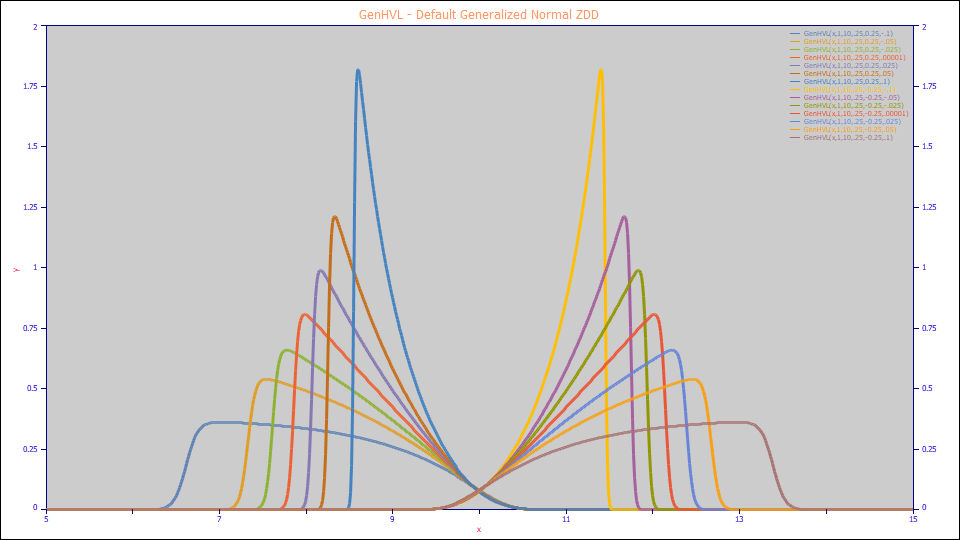
GenHVL - Default Generalized Normal ZDD
The GenHVL model:
a0 = Area
a1 = Center (as mean of asymmetric peak)
a2 = Width (SD of underlying Gaussian ZDD)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = ZDD asymmetry ( -1 > a4 > 1 )
Built in model: GenHVL
User-defined peaks and view functions: GenHVL(x,a0,a1,a2,a3,a4)
GenHVL[Z] - [Z] Generalized Normal ZDD
The GenHVL[Z] model:
a0 = Area
a1 = Center (as mean of generalized normal ZDD)
a2 = Width (SD of underlying Gaussian)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = ZDD asymmetry ( -1 > a4 > 1 )
Built in model: GenHVL[Z]
User-defined peaks and view functions: GenHVL[Z](x,a0,a1,a2,a3,a4)
![v5_GenHVL[Z].png](v5_GenHVL[Z].png)
GenHVL[Y] - [Y] Generalized Error Model ZDD
The GenHVL[Y] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying normal ZDD)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = Power n in exp(-zn) decay ( .25 > a4 > 4 ) adjusts kurtosis (fourth moment)
a5 = ZDD asymmetry ( -1 > a5 > 1 ), adjusts skew (third moment)
Built in model: GenHVL[Y]
User-defined peaks and view functions: GenHVL[Y](x,a0,a1,a2,a3,a4,a5)
![v5_GenHVL[Y].png](v5_GenHVL[Y].png)
GenHVL[T] - [T] Generalized Student's t ZDD
The GenHVL[T] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying normal ZDD)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = Student's t tailing, the nu or DOF ( 1 > a4 > 1,000,000 ) (fourth moment)
a5 = ZDD asymmetry ( -1 > a5 > 1 ), adjusts skew (third moment)
Built in model: GenHVL[T]
User-defined peaks and view functions: GenHVL[T](x,a0,a1,a2,a3,a4,a5)
![v5_GenHVL[T].png](v5_GenHVL[T].png)
GenHVL[V] - [V] Generalized Error Model ZDD
The GenHVL[V] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying normal ZDD)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = The GMG half-Gaussian convolution width, adjusts skew (third moment)
a5 = ZDD asymmetry ( -1 > a5 > 1 ), adjusts skew (third moment)
Built in model: GenHVL[V]
User-defined peaks and view functions: GenHVL[V](x,a0,a1,a2,a3,a4,a5)
![v5_GenHVL[V].png](v5_GenHVL[V].png)
GenHVL[G] - [G] Half-Gaussian Modified Gaussian (GMG)
The GenHVL[G] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying Gaussian)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = The GMG half-Gaussian convolution width
Built in model: GenHVL[G]
User-defined peaks and view functions: GenHVL[G](x,a0,a1,a2,a3,a4)
![v5_GenHVL[G].png](v5_GenHVL[G].png)
GenHVL[E] - [E] Exponentially-Modified Gaussian (EMG)
The GenHVL[E] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying Gaussian)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = The EMG half-Gaussian convolution width
Built in model: GenHVL[E]
User-defined peaks and view functions: GenHVL[E](x,a0,a1,a2,a3,a4)
![v5_GenHVL[E].png](v5_GenHVL[E].png)
GenHVL[K] - [K] Generalized ZDD
The GenHVL[K] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying normal ZDD)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = The fractional order of the decay from the apex of the peak upward
a5 = The fractional order of the rise to the apex
Built in model: GenHVL[K]
User-defined peaks and view functions: GenHVL[K](x,a0,a1,a2,a3,a4,a5)
![v5_GenHVL[K].png](v5_GenHVL[K].png)
GenHVL[Q] - [Q] Error Model (Symmetric)
The GenHVL[Q] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying Gaussian)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = ZDD power of tailing ( 0.25 > a4 > 4 )
Built in model: GenHVL[Q]
User-defined peaks and view functions: GenHVL[Q](x,a0,a1,a2,a3,a4)
![v5_GenHVL[Q].png](v5_GenHVL[Q].png)
GenHVL[S] - [S] Student's t (Symmetric)
The GenHVL[S] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying Gaussian)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = Student's t tailing, the nu or DOF ( 1 > a4 > 1,000,000 )
Built in model: GenHVL[S]
User-defined peaks and view functions: GenHVL[S](x,a0,a1,a2,a3,a4)
![v5_GenHVL[S].png](v5_GenHVL[S].png)
GenHVL[YpE] - [YpE] Generalized Laplace ZDD
The GenHVL[YpE] model:
a0 = Area
a1 = Center (as mean of underlying normal ZDD)
a2 = Width (SD of underlying normal ZDD)
a3 = HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = ZDD asymmetry ( -1 > a4
> 1 ), adjusts skew (third moment)
Built in model: GenHVL[YpE]
User-defined peaks and view functions: GenHVL[YpE](x,a0,a1,a2,a3,a4)
![v5_GenHVL[D].png](v5_GenHVL[D].png)
GenHVL[Yp] - [Yp] Generalized Error Model ZDD
The GenHVL[Yp] model model is identical to GenHVL[Y] model, but uses a separate starting estimate algorithm which assumes high overload preparatory peak shapes.
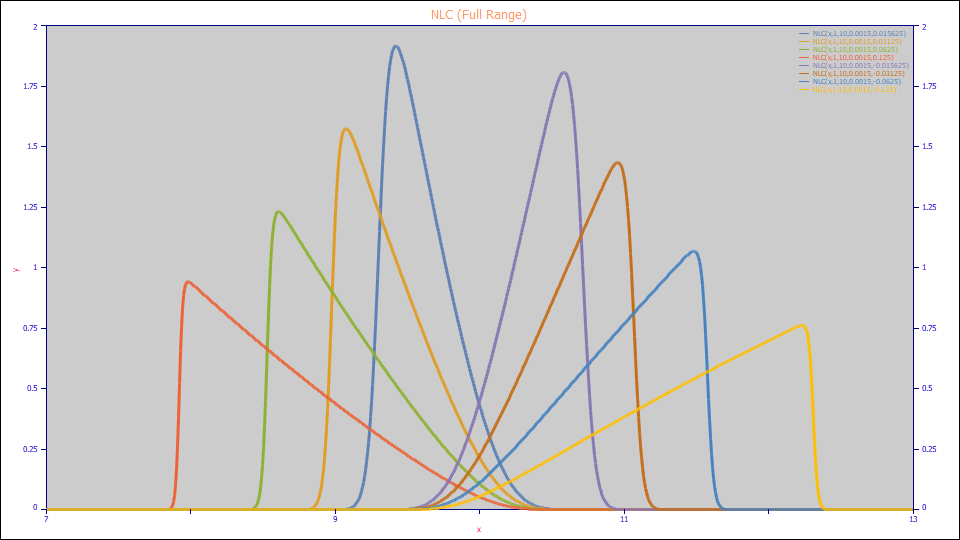
NLC (Wade-Thomas Non-Linear Chromatography)
The NLC (full range) model:
7
a0 = Area
a1 = Center (as mean of asymmetric peak)
a2 = Width (NLC/Giddings kinetic time constant)
a3 = NLC chromatographic distortion ( -1 > a3 > 1 )
Built in model: NLC
User-defined peaks and view functions: NLC(x,a0,a1,a2,a3)
GenNLC[Z] - [Z] ZDD
The GenNLC[Z] is derived using the relationships of equivalence between the GenHVL and GenNLC models:
To convert the GenHVL[Z] to the GenNLC[Z], the a2 is converted to a Giddings kinetic time constant, the a4 is converted to a Gidding's indexed asymmetry.
a0 = Area
a1 = Center (as mean of generalized normal ZDD)
a2 = Kinetic Width (Giddings time constant of ZDD)
a3 = NLC/HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = NLC indexed asymmetry ( -10 > a4 > 10 ) a4=0.5 NLC (Giddings)
Built in model: GenHVL[Z]
User-defined peaks and view functions: GenHVL[Z](x,a0,a1,a2,a3,a4)
![v5_gennlc[z].png](v5_gennlc[z].png)
GenNLC[Y] - [Y] ZDD
The GenNLC[Y] model is derived using the relationships of equivalence between the GenHVL and GenNLC models:
To convert the GenHVL[Y] to the GenNLC[Y], the a2 is transformed to a Giddings kinetic time constant, the a5 is transformed to a Gidding's indexed asymmetry.
a0 = Area
a1 = Center (as mean of generalized normal ZDD)
a2 = Kinetic Width (Giddings time constant of ZDD)
a3 = NLC/HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = Power n in exp(-zn) decay ( .25 > a4 > 4 ), adjusts kurtosis (fourth moment)
a5 = NLC indexed asymmetry ( -10 > a5 > 10 ) a5=0.5 NLC (Giddings), adjusts skew (third moment)
Built in model: GenNLC[Y]
User-defined peaks and view functions: GenNLC[Y](x,a0,a1,a2,a3,a4,a5)
![v5_GenNLC[Y].png](v5_GenNLC[Y].png)
GenNLC[T] - [T] ZDD
The GenNLC[T] model is derived using the relationships of equivalence between the GenHVL and GenNLC models:
To convert the GenHVL[T] to the GenNLC[T], the a2 is transformed to a Giddings kinetic time constant, the a5 is transformed to a Gidding's indexed asymmetry.
a0 = Area
a1 = Center (as mean of generalized normal ZDD)
a2 = Kinetic Width (Giddings time constant of ZDD)
a3 = NLC/HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = Student's t tailing, the nu or DOF ( 1 > a4 > 1,000,000 ) (fourth moment)
a5 = NLC indexed asymmetry ( -10 > a5 > 10 ) a5=0.5 NLC (Giddings), adjusts skew (third moment)
Built in model: GenNLC[T]
User-defined peaks and view functions: GenNLC[T](x,a0,a1,a2,a3,a4,a5)
![v5_GenNLC[T].png](v5_GenNLC[T].png)
GenNLC[V] - [V] ZDD
The GenNLC[V] model is derived using the relationships of equivalence between the GenHVL and GenNLC models:
To convert the GenHVL[V] to the GenNLC[V], the a2 is transformed to a Giddings kinetic time constant, the a5 is transformed to a Gidding's indexed asymmetry.
a0 = Area
a1 = Center (as mean of generalized normal ZDD)
a2 = Kinetic Width (Giddings time constant of ZDD)
a3 = NLC/HVL Chromatographic distortion ( -1 > a3 > 1 )
a4 = The GMG half-Gaussian convolution width, adjusts skew (third moment)
a5 = NLC indexed asymmetry ( -10 > a5 > 10 ) a5=0.5 NLC (Giddings), adjusts skew (third moment)
Built in model: GenHVL[V]
User-defined peaks and view functions: GenHVL[V](x,a0,a1,a2,a3,a4,a5)
![v5_GenNLC[V].png](v5_GenNLC[V].png)
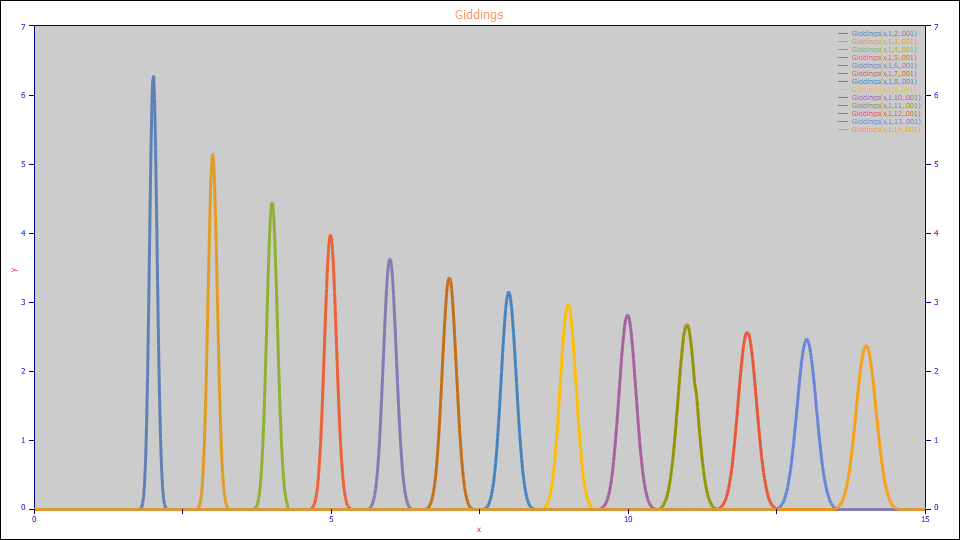
The Giddings Model
The Giddings model:
a0 = Area
a1 = Center (as mean of asymmetric peak)
a2 = Kinetic Width (as time constant)
Built in model: Giddings
User-defined peaks and view functions: Giddings(x,a0,a1,a2)
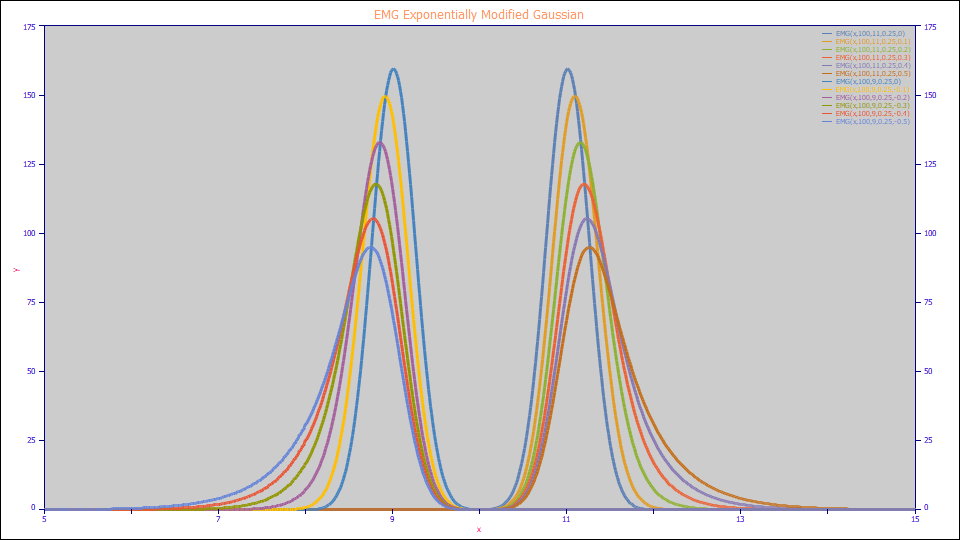
EMG Exponentially Modified Gaussian
The EMG Model:
a0 = Area
a1 = Center (as mean of Gaussian peak that is convolved by the a3 exponential)
a2 = Width (SD of the Gaussian peak that is convolved by the a3 exponential)
a3 = The first order kinetic or exponential decay width convolving the Gaussian (can be negative to model fronted peaks)
Built in model: EMG
User-defined peaks and view functions: EMG(x,a0,a1,a2,a3)
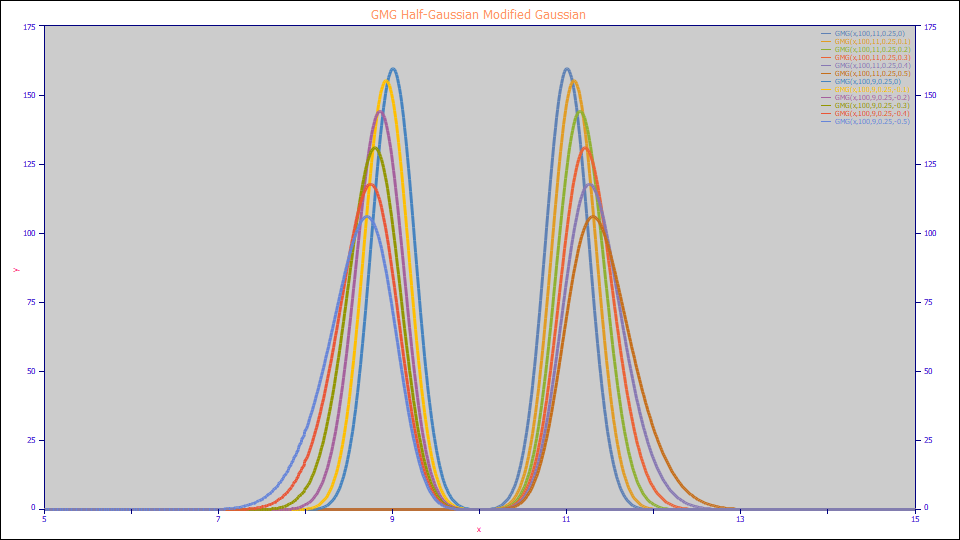
GMG Half-Gaussian Modified Gaussian
The GMG Model:
a0 = Area
a1 = Center (as mean of Gaussian peak that is convolved by the a3 half-Gaussian)
a2 = Width (SD of the Gaussian peak that is convolved by the a3 half-Gaussian)
a3 = The first order probabilisitic half-Gaussian width convolving the Gaussian (can be negative to model fronted peaks)
Built in model: GMG
User-defined peaks and view functions: GMG(x,a0,a1,a2,a3)
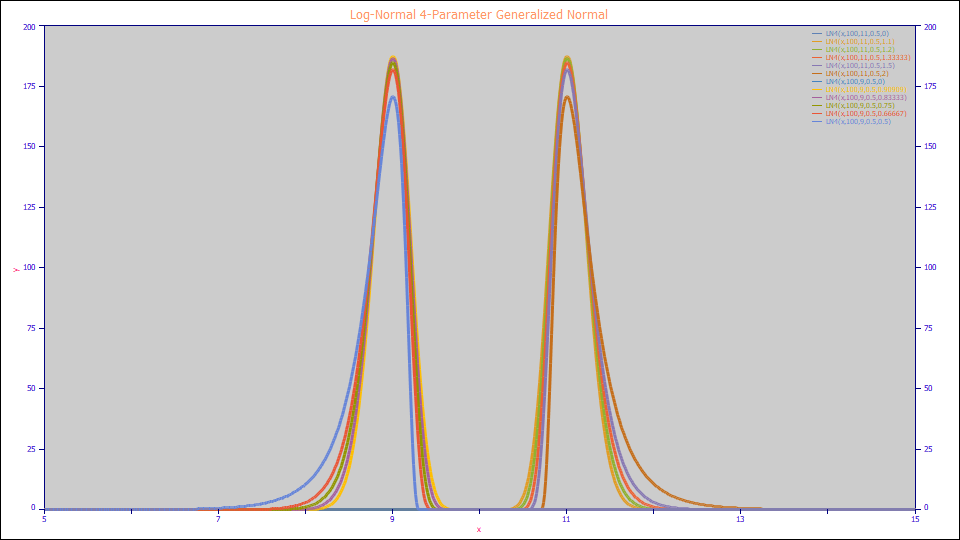
Log-Normal 4-Parameter Generalized Normal
The LN4 Model:
a0 = Area
a1 = Center (mode of asymmetric peak)
a2 = Half-Height Width (FWHM)
a3 = Half-Height Asymmetry Ratio (Asym50)
Built in model: LN4 or GenNorm[cwa]
User-defined peaks and view functions: LN4(x,a0,a1,a2,a3) or GenNorm[cwa](x,a0,a1,a2,a3)


 |