PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
Equivalence GenHVL and GenNLC
Common Chromatographic Distortion Operator
In the topic ZDD Concepts, the derivation of the GenHVL and GenNLC models is given. Here we revisit the basic Generalized HVL and NLC templates with respect to equivalence in modeling this specific shapes of chromatographic peaks.
The HVL and NLC models cannot produce the same chromatographic peak shapes. The HVL assumes a Gaussian zero-distortion density (ZDD) and the NLC assumes a Giddings ZDD or infinite dilution density. The HVL is computes a theoretical diffusion width, the NLC a theoretical adsorption-desorption kinetic time constant.
If a generalization of the HVL model is capable of exactly modeling an NLC shape, and a generalization of the NLC model is capable of exactly modeling the HVL shape, there will exist a set of relationships between the parameters of these models that will allow a model based on a diffusion width, such as the GenHVL, to be converted or translated to a model based on a kinetic time constant, such as the GenNLC.
We feel it is highly significant that there exists a common chromatographic distortion between the only diffusion model we found to be consistently effective in the chromatographic modeling of widely varying concentrations, the HVL, and the only kinetic model we saw as consistently effective, the Wade-Thomas NLC. It suggests the very real possibility of a general chromatographic model that can manage a wide range of kinetic, diffusion, or lumped contribution effects in chromatographic peak band broadening, and this is accomplished very simply, using only a common chromatographic operator and a specific density assumption.
One simply extracts that parametric broadening as a pure diffusion width, or a pure first order kinetic time constant, or as both as a consequence of this equivalence, but only after removing by fitting the non-idealities that would otherwise distort those parameters. While the parameters of that nonideality may also be of importance, the primary idea of equivalence is to extract the a2 band broadening had the eluted peak been a perfect HVL with its Gaussian density assumption at infinite dilution or a perfect NLC with its Giddings density assumption at infinite dilution.
The Generalized HVL and NLC Templates
In the ZDD concepts topic, we defined a simple HVL generalized template that accepts any zero-distortion density:
A simple NLC density generalized template was derived as follows:
At this point, we again note our reparameterization of the NLC a2 as a time constant rather than a rate constant, making it identical to the HVL as a statistical scale parameter.
To regenerate the HVL, the HVL Template is used with the normal PDF and CDF replacing the Density and Cumulative in the formula. To regenerate the NLC, the NLC template is used with the Giddings PDF and CDF complement replacing the Density and RevCumulative in the formula.
What we nowhere expected to find, and which no one to our knowledge had yet discovered, is that one can regenerate the HVL shape by using the NLC template with the Gaussian PDF and CDF complement, and one can regenerate the NLC shape by using the HVL template with the Giddings PDF and CDF.
The obvious inference is that the insertion of a common PDF, CDF, and CDFc into the two templates will result in the capability of generating identical peak shapes, ideally one parameterization designed to model a diffusion-based band broadening, the other a kinetic one. Although the parameters will be different, the fits realizable in mathematical modeling are identical.
The a3 in all GenHVL and GenNLC models is thus a common chromatographic distortion which we treat as a pure mathematical distortion operator. In fact, we reparameterized the GenNLC models to have the same exact a3 distortion values as the GenHVL, a simple matter since these differed only by a scalar factor of two.
Interpreting a2 Widths
In the course of our work, J. L. Wade, the original author of the NLC model, stressed to us that a2 was a lumped kinetic parameter, and likely included kinetics that did not specifically pertain to the adsorption process his model specifically mapped. He also pointed out those differences which were well described in the Horvath and Lin papers, and the Arnold papers, of the 1980s, as well as his own subsequent one, to highlight the profoundly different kinetic processes that would be 'lumped' into this singular kinetic a2 in his model, or into any model that sought to specify a single theoretical kinetic width for the complex multiple processes occurring in chromatographic band broadening.
We acknowledge that the NLC specific a2 is based on a specific set of assumptions, just as the a2 of the HVL assumes a pure Gaussian diffusion at infinite dilution. What PeakLab adds, however, is the capacity to go beyond these assumptions in fitting densities that allow for significant deviations from the specific HVL and NLC theory. We can model the real-world zero-distortion density by using generalizations that not only accommodate the pure HVL and NLC assumptions, but potentially every non-ideality that can be represented in some form of a statistical density model. Again, note the simple templates above. The whole of the band broadening will ultimately be represented in the density or ZDD used. It is why ZDDs are so critical in understanding the innovations PeakLab brings to fitting chromatographic peaks.
The GenHVL and GenNLC are not empirical models. They are statistical extensions or generalizations of sound theoretical models. A good statistical fitting and deconvolution should remove, isolate, quantify, or otherwise characterize the non-idealities which are not consistent with the specific density assumptions of the NLC and HVL models. In a GenHVL or GenNLC fit, these deviations from ideality are actually isolated and quantified in the higher moment fitting that occurs in the ZDD component of generalized models. Or, to put it another way, we perform a mathematical separation where only like broadening is included the a2 primary width term, and a single a4 density non-ideality term, shared across all peaks, will account and separate that broadening that is not compatible with the simplest of the diffusion assumptions, a Gaussian and its SD width, or a first order kinetic process, a Giddings with its specific a2 time constant.
We accept, at infinite dilution, the real-world diffusion will be non-Gaussian, the real-world kinetics non-Giddings, but the generalized densities account those differences and separate them, reporting a true Gaussian width for that portion of the broadening where that assumption is valid, and a true dimensionless time constant based on first order kinetics for the broadening where that particular assumption applies. Further, the modeling elegantly quantifies the measure of deviation from that nonideality.
When the same identical density (ZDD) is used in both generalized models, it is possible to extract the pure HVL width, the Gaussian a2 diffusion at infinite dilution, and the pure NLC width, the pure first order kinetics of the Giddings a2, even though the information being partitioned or regarded as part of the nonideality may be quite different. This is the basic premise of GenHVL and GenNLC equivalence.
The statistical model is shape-equivalent. The fitted peak shape will be identical when this non-ideality parameter is not shared across multiple peaks. Note that we are not equating diffusion and kinetics or any aspect of the underlying mechanisms of broadening. We are only noting an interconversion arising from two different parametrizations of a single model.
Fitting Higher Moments
We do not suggest the Wade-Thomas (Giddings) a2 is an ultimate representation of the kinetics for any chromatographic peak where kinetics are viewed as the source of band broadening, any more than we suggest the HVL (Gaussian) a2 is an ultimate representation of the diffusion when it is the source of the broadening in chromatographic peaks. What we do suggest is that these two values represent the simplest of these two specific characterizations of peak broadening in chromatography. Anything beyond a Gaussian for diffusion will be appreciably more complex. Anything beyond the Giddings type of kinetics in the Wade-Thomas NLC will be immensely complex, and the Wade-Thomas model itself stretched the bounds of computational modeling when it was first published.
The generalized models allow us to have 'one' picture, and we believe a very elegant, simple, and highly favorable one with respect to mathematical modeling, of the overall kinetics of the band-broadening process. They also allow us to have 'one' very different picture of the diffusive band broadening, equally elegant, simple, and ideal for mathematically modeling chromatographic peaks.
Specifically we fit the higher moments of the ZDD in the GenHVL models and we do so in the context of deriving a singular a2 diffusion width, based on the HVL Template and this Gaussian assumption. Looking closely at template, we make only one statistical assumption, the same one upon which the HVL is fundamentally based, that the diffusion is normally distributed, and the a2 is that normal distribution width. The a3 is this common chromatographic distortion operator shared with the NLC.
Similarly we fit the higher moments of the ZDD in the GenNLC models and we do so in the context of deriving a singular a2 overall kinetic time constant, based on the NLC Template. Looking closely at its template, we are again making only one statistical assumption, the same one upon which the NLC reduces at infinite dilution, that the essential ZDD to be retained in the a2 is the Giddings kinetic term. Again, the a3 is this same common chromatographic distortion operator shared with the HVL.
Using nonlinear fitting to separate nonidealities works only when the different effects can be mathematically isolated in the iterative optimization procedures. Multiple Gaussian effects are likely to be lumped together in the GenHVL's a2; multiple first order kinetic effects are likely to be lumped together in the GenNLC's a2. Only those nonidealities separable in the fitting as non-Gaussian or non-first-order can be accurately isolated. For such fitting, quality data, a high S/N, is essential.
Models of Equivalence
For a GenNLC to be constructed two items are necessary. First, the generalization in the ZDD must be able to accurately model all Giddings shapes, and to a high degree of accuracy. Second, since the Giddings shape is location dependent, a mathematical relationship must account this dependency so that a single asymmetry parameter will always produce the Giddings ZDD (and thus the NLC with the applied a3 chromatographic distortion to the Giddings ZDD), irrespective of the specific a1 of the peak. In the statistical sciences there are three generalized normals widely used:
1. The asymmetric generalized normal, the default and [Z] ZDDs in the program
2. The GMG or Skew Normal model, the [G] ZDD in the program.
3. The Error model, the [Q] ZDD in the program
The [Q] can produce only symmetric peaks (as is true of the [S] Student's t ZDD). The [G] can somewhat approximate the Giddings, but the asymmetric generalized normal ZDDs do so to a high level of accuracy, and they offer very elegant and simple equivalence relationships. As such the GenNLC models only exist where the Giddings ZDD (and thus a pure NLC), is a specialization at a fixed single a4 (or a5) asymmetry parameter, irrespective of the a1 location or center of the peak. For PeakLab, this means GenNLCs are only constructed for the default and [Z] ZDD models, and for the [Y] , [T] , and [V] densities.
We note that the only difference between the default ZDD and the [Z] is that the PeakLab default parameterizes a1 as the mean or centroid of the peak, and [Z] the standard statistical definition where a1 is the median.
GenHVL and GenNLC Equivalence
To report the equivalence of the a2 width and what is usually an a4 non-ideality asymmetry, we will use the A0-A4 nomenclature for the parameters of the GenNLC parameters and a0-a4 for the GenHVL parameters. Note that a4 and A4 will be a5 and A5 in the [Y], [T], and [V] ZDD models.
This generalized ZDD which supports both the HVL-based and NLC-based chromatographic models requires only these simple identities of equivalence. The a0 area, a1 centroid, and a3 chromatographic distortions are identical. Only the a2 width, and the statistical asymmetry parameter that addresses the nonideality in the density, are adjusted.
In the Numeric Summary, the Equivalent Parameters section will use these relationships to convert between GenHVL and GenNLC parametrizations. Using this report option, you can view these simple diffusion and kinetic widths, absent the nonidealities which were separable by nonlinear modeling.
Why GenNLC Models are Necessary
If it is possible to convert easily between the two models, why not simply fit only GenHVLs, and perform the conversions above? The answer lies in the sharing of what is usually this a4 asymmetry parameter. In the GenHVL, the a4 is a pure statistical asymmetry. The higher the a4 value, the higher the asymmetry of the ZDD in the model. The peaks are indexed to the zero asymmetry of the Gaussian in the HVL.
For the GenNLC, the Giddings asymmetry varies with the a1 position. If a set of pure NLC peaks are independently fitted to the GenHVL, the a4 asymmetries will not be constant across the different peaks because of this locational dependency. In order to render the GenNLC indexed to the Giddings, a separate GenNLC must be fitted which maintains a constant a4 asymmetry for the GenNLC's ZDD. If that ZDD is the Giddings, meaning each peak is a pure NLC, the a4 of the GenNLC will always be 1/2. The GenNLC is thus indexed to Giddings ZDD, a constant a4=0.5 for a pure NLC peak, regardless of where it is positionally.
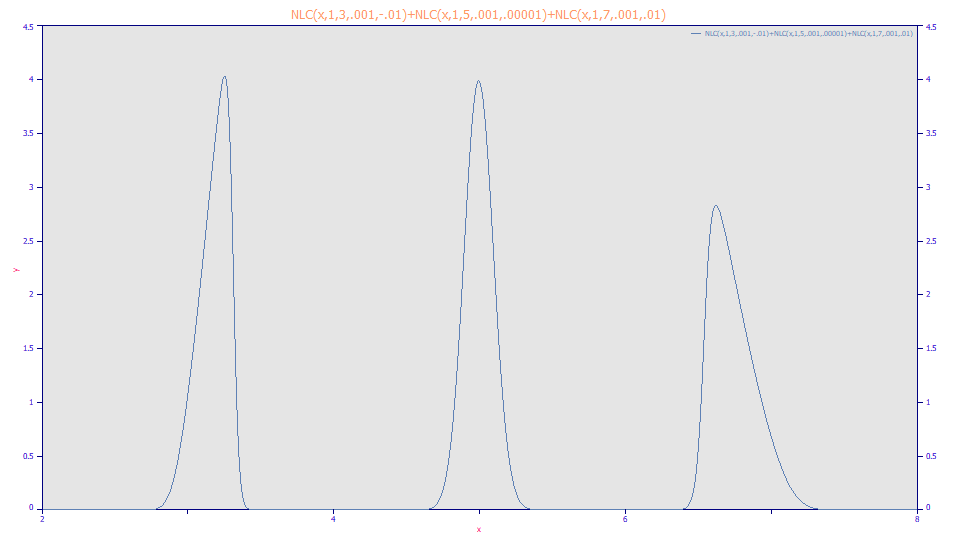
We have created three pure NLC peaks, giving the first a fronted shape, the second essentially symmetric, and the third a tailed shape, each with the same a2 kinetic time constant. Let's fit these first to the GenNLC with a4 shared across all peaks:
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
1.00000000 1.00000000 6.6858e-15 1.8852e+31 0.00000000
Peak Type a0 a1 a2 a3 a4
1 GenNLC 1.00000000 3.00000000 0.00100000 -0.0200000 0.50000000
2 GenNLC 1.00000000 5.00000000 0.00100000 2e-5 0.50000000
3 GenNLC 1.00000000 7.00000000 0.00100000 0.02000000 0.50000000
Equivalent Parameters
Peak Type a0 a1 a2 a3 a4
1 GenHVL 1.00000000 3.00000000 0.07745967 -0.0200000 0.01290994
2 GenHVL 1.00000000 5.00000000 0.10000000 2e-5 0.01000000
3 GenHVL 1.00000000 7.00000000 0.11832160 0.02000000 0.00845154
Measured Values
Peak Type Amplitude Center FWHM Asym50 FW Base Asym10
1 GenNLC 4.03560334 3.25634702 0.23602109 0.36415363 0.47524715 0.30899960
2 GenNLC 3.98977994 4.99810117 0.23545299 1.01314665 0.47134237 1.02334341
3 GenNLC 2.83232374 6.61807038 0.33147623 2.79451428 0.69086881 3.42867327
Peak Type Area % Area Mean StdDev Skewness Kurtosis
1 GenNLC 1.00000000 33.3333333 3.17698891 0.10587272 -0.6351959 2.92081263
2 GenNLC 1.00000000 33.3333333 4.99971790 0.10000545 0.03141248 3.00172781
3 GenNLC 1.00000000 33.3333333 6.73705501 0.15561907 0.70114218 3.05677794
All Total 3.00000000 100.000000
Note that the peaks are perfectly fitted. There is an increasing FWHM with location despite having a constant a2 kinetic width term. Note also that the a4 is a perfect 0.5 for all three peaks, indicating a pure NLC (a pure Giddings ZDD). Note also that the equivalent GenHVL parameters show an increasing a2 instead of a constant value, and an a4 statistical asymmetry that also decreases considerably with a1 location.
Now let us fit the GenHVL, again with a4 a single fitted parameter shared across the three peaks:
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999888 0.99999887 0.00106667 7.4063e+8 1.12381843
Peak Type a0 a1 a2 a3 a4
1 GenHVL 1.00032218 2.99988459 0.07798367 -0.0201522 0.01121410
2 GenHVL 0.99999882 4.99957246 0.09999502 -1.184e-5 0.01121410
3 GenHVL 1.00053127 7.00027514 0.11958643 0.02023303 0.01121410
Equivalent Parameters
Peak Type a0 a1 a2 a3 a4
1 GenNLC 1.00032218 2.99988459 0.00101361 -0.0201522 0.43138520
2 GenNLC 0.99999882 4.99957246 0.00099999 -1.184e-5 0.56068491
3 GenNLC 1.00053127 7.00027514 0.00102145 0.02023303 0.65644385
Measured Values
Peak Type Amplitude Center FWHM Asym50 FW Base Asym10
1 GenHVL 4.03847166 3.25628056 0.23563768 0.36514873 0.47578760 0.30895600
2 GenHVL 3.98976154 4.99812663 0.23545483 1.01251933 0.47133942 1.02337770
3 GenHVL 2.83524745 6.61824121 0.33064669 2.78132352 0.69207943 3.42791279
Peak Type Area % Area Mean StdDev Skewness Kurtosis
1 GenHVL 1.00032218 33.3346027 3.17689067 0.10605143 -0.6396915 2.93206083
2 GenHVL 0.99999882 33.3238269 4.99973948 0.10000588 0.03280949 3.00192836
3 GenHVL 1.00053127 33.3415704 6.73729673 0.15606415 0.70862146 3.07701793
All Total 3.00085227 100.000000
First note that the fit is no longer perfect. The shared a4 ZDD statistical asymmetry makes it impossible to fit three perfect NLC peaks. There is error, albeit a small one. Note that a2, as a diffusion width independent of location, tracks the FWHM. Note also the equivalent parameters. The equivalent GenNLC a2 are close to the constant value of the NLC's used to generate the data, the a4 are the vicinity of 0.5, but there is no indication of a pure NLC anywhere in the fit. And there is none; the shared a4 of an asymmetry indexed to a symmetric ZDD preventing such.
We will make one more instructive fit, allowing the a4 asymmetry of the GenHVL to vary on a per-peak basis (it will be independently fitted for each peak).
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
1.00000000 1.00000000 6.7046e-15 1.6068e+31 0.00000000
Peak Type a0 a1 a2 a3 a4
1 GenHVL 1.00000000 3.00000000 0.07745967 -0.0200000 0.01290994
2 GenHVL 1.00000000 5.00000000 0.10000000 2e-5 0.01000000
3 GenHVL 1.00000000 7.00000000 0.11832160 0.02000000 0.00845154
Equivalent Parameters
Peak Type a0 a1 a2 a3 a4
1 GenNLC 1.00000000 3.00000000 0.00100000 -0.0200000 0.50000000
2 GenNLC 1.00000000 5.00000000 0.00100000 2e-5 0.50000000
3 GenNLC 1.00000000 7.00000000 0.00100000 0.02000000 0.50000000
Measured Values
Peak Type Amplitude Center FWHM Asym50 FW Base Asym10
1 GenHVL 4.03560334 3.25634702 0.23602109 0.36415363 0.47524715 0.30899960
2 GenHVL 3.98977994 4.99810117 0.23545299 1.01314665 0.47134237 1.02334341
3 GenHVL 2.83232374 6.61807038 0.33147623 2.79451428 0.69086881 3.42867327
Peak Type Area % Area Mean StdDev Skewness Kurtosis
1 GenHVL 1.00000000 33.3333333 3.17698891 0.10587272 -0.6351959 2.92081263
2 GenHVL 1.00000000 33.3333333 4.99971790 0.10000545 0.03141248 3.00172781
3 GenHVL 1.00000000 33.3333333 6.73705501 0.15561907 0.70114218 3.05677794
All Total 3.00000000 100.000000
As a validation of statistical equivalence on an individual peak basis, note that three NLC peaks are now perfectly fitted by the GenHVL model. We now have in the Equivalent Parameters section the parameter values that we realized in directly fitting the GenNLC with a shared a4. Note the a4 values in the GenHVL, indexed to an absolute asymmetry, that were necessary to produce the constant a4 GenNLC.
Sharing this a4 parameter is recommended. It may well be necessary, even if peaks are cleanly separated, if the S/N is limited or the IRF instrumental influence is inaccurately managed. You will thus want to use the GenNLC if you wish to fit widths indicative of first order kinetics. You will want to use the GenHVL if you wish fitted widths indicative of a positionally-independent band broadening. The Equivalent Parameters will give you a good picture of the kinetics when you fit the GenHVL, and a good picture of a diffusion-type broadening when you fit the GenNLC.


 |