PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
User-Defined Peaks
![]() You may install up to sixty non-linear User-Defined Functions (UDFs) into PeakLab's peak set at any given
time. This User-Defined Functions option is used as the control center for entering, editing,and saving
UDFs. From this UDF dialog, you can work with individual functions or with UDF libraries.
You may install up to sixty non-linear User-Defined Functions (UDFs) into PeakLab's peak set at any given
time. This User-Defined Functions option is used as the control center for entering, editing,and saving
UDFs. From this UDF dialog, you can work with individual functions or with UDF libraries.
When a user function library is used within fits, or simply loaded, the PeakLab data file will also save those user functions. These will remain available within the PeakLab data file, but any revisions to an existing user function which was used in fits will invalidate those specific fits.
The current UDFs are also automatically saved upon program exit as a UDF library. The configuration file PF2.CFG, is actually this UDF library. This file is automatically written upon exit, but an existing file is only overwritten when a UDF has been installed in the current session. When this Last Session UDFs item is selected, these UDFs are loaded, validated, and compiled.
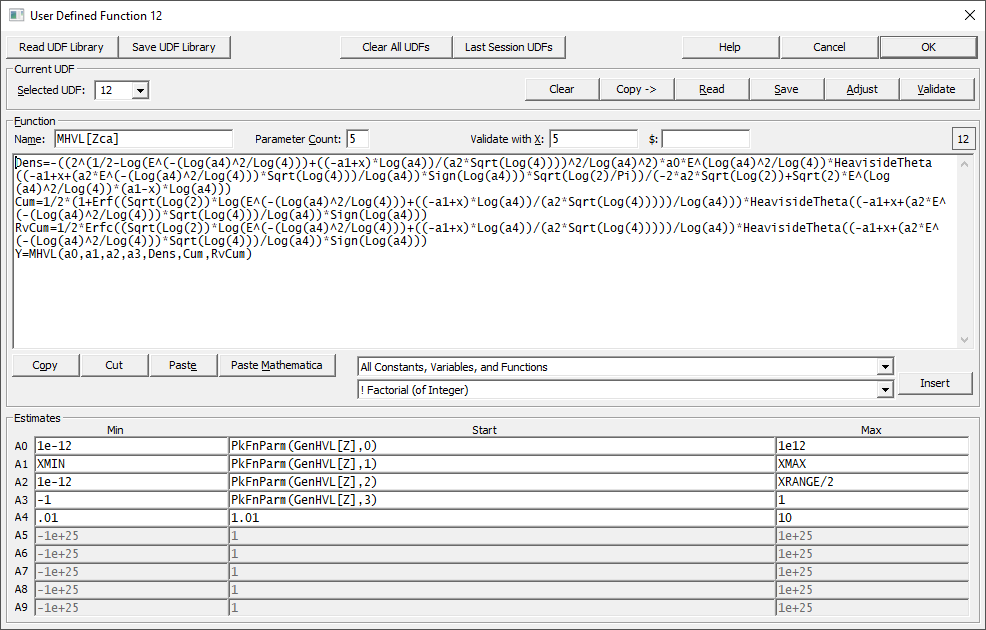
User Defined Functions as "User Defined Peaks"
In PeakLab, the user functions have become full user-defined peaks which can be used interchangeably with built-in models. The sophistication of the models can also be much greater and there is now Maple and Mathematica syntax support for entering such peak models. The only significant change to enable a user-defined model to become a fully flexible peak with the capabilities of built in models is that you must now also specify formulas for the starting estimates.
If you are familiar with the UDFs of prior versions of the product, there are a number of important changes. Most functions in the parser are now case-sensitive, although many of the more common mathematical functions are still case insensitive to support direct import of Maple and Mathematica expressions. The function list has also grown and the help for functions is now included in the dialog.
We strongly suggest you build your models as we do with parameters a0, a1, a2, and a3 tracking moments 0, 1, 2, 3. We suggest that you parameterize the user model parameter associations so that a0 is an area or amplitude, a1 is the center or location, a2 is related to the peak width, and a3 is related to the skew or third moment (for chromatography, asymmetry or distortion). We have found that this simplifies the authoring of such user-defined models. In most cases, you should be able set a4 and higher with a fixed value for a starting estimate, as we did in the above example.
PeakLab User Defined Functions
You can add up to 60 user-defined peak models to a UDF library, and all of these can be imported and made available to any data set. You can have as many UDF libraries as you like, but only one can be loaded at any given time.
Each user function will be compiled internally to run at close to the speed of built-in functions. The slower speed of UDF fits arises from the need to compute numeric derivatives, which can require many computations of the function. Built-in functions use analytic derivatives that use appreciably less computation and contain no computational error.
The main user function expression can contain up to nine separate variables or expressions which are then used in the final Y= user function definition.
The main user function expression can be up to 8192 characters in length. The Start value formulas can each be up to 2048 characters. In the above graphic, the main user function expression consists of just over 700 characters. You should not run into any limits with any peak model you wish to explore.
If you wish to use PeakLab as a 'peak laboratory' for experimenting with new chromatographic models, you will probably in the course of such work create a model similar to that in the example above where a PDF (a probability density function), a CDF (a cumulative distribution function), and a CDFc (a cumulative distribution complement) is used with a generalized HVL or NLC model template function. We created a huge number of such models in the course of PeakLab's development. Many were tested as user-defined peaks before being coded as built-in models.
Active UDFs are available for placement in PeakLab's three fitting options. Note that PeakLab's UDFs cannot be used to fit data directly. Rather are they assumed to describe components, usually individual peaks, that are placed during the prefit peak-placement process.
Within the fitting procedures, a UDF is directly selected and it will place based on the formulas for the starting estimates.
To simplify the fitting of IRF convolution integrals of user functions, each user function you create will also be automatically created with an <e>, <ge>, and <e2> IRF. Any peak model you create will thus automatically be capable of fitting these three IRFs which are far and away those we have found most useful in chromatographic modeling. So long as you can treat the instrumental distortion with one of these principal instrumental/system distortion models, no additional effort is needed. You do not need to create an convolution integral UDF.
Note also that UDFs need not be peak functions. They can be unusual baselines, transitions, or any other function desired to describe any unusual feature in the data.
A PeakLab UDF contains all information necessary to describe and initially place the function, including the function name, the parameter count, the function's formula, the starting estimate formulas, and constraints for each parameter.
Viewing a User-Defined Peak
If you create a user function and you wish to explore it graphically, you can use the View Function(X) option with the UDF1P(n,x,a0) , UDF2P(n,x,a0,a1), ... UDF8p(n,x,a0,a1,a2,a3,a4,a5,a6,a7) parser functions. Please be sure you have entered and exited the user functions dialog to ensure the functions have been compiled and are available to the program.
The View Function(X) option allows you to export data sets to disk which can be subsequently imported and fitted. Since the View Function(X) rendering of user-defined functions uses your own values for the parameters, not the starting estimates formulas, these do not have to be established to view the function.
If you are uncertain of how to generate starting estimates for your user-defined peak model, you can import data consisting of a sum of a couple of user-defined peaks with known parameters and run a Model Experiment with various built-in models to see if any generate similar parameters.
Starting Estimate Formulas
In order to place multiple peaks with different areas, positions, widths, and shapes, it is essential the starting estimate formulas produce reasonable estimates based on the peak which is automatically being placed by the program's algorithms. The easiest way to do this is to 'borrow' the programs's own estimation routines for the parameters of its built-in functions
PkFnParm()
This is the main function we use for starting estimates. The formula PkFnParm(GenHVL[Z],0) for the A0 parameter borrows the program's a0 parameter estimate for a GenHVL[Z] peak model. With PkFnParm(), you don't need to know anything about how to generate a starting estimate, only that a given built-in model generates a parameter reasonably close to your model.
If you are exploring different ZDD densities using the common chromatographic distortion model in your user-defined peak, virtually any model using a Gaussian SD type of a2 width should should be fine with PkFnParm(GenHVL,0), PkFnParm(GenHVL,1), PkFnParm(GenHVL,2), PkFnParm(GenHVL,3), for the A0-A3 estimates as in the example above. If your user-defined peak is of the Giddings' type with first order kinetics reflected in a2, you should be fine with PkFnParm(GenNLC,0), PkFnParm(GenNLC,1), PkFnParm(GenNLC,2), PkFnParm(GenNLC,3), for the A0-A3 estimates. If your model uses a rate instead of a time constant, you may need to invert and possibly scale the PkFnParm(GenNLC,2) kinetic a2 value to make it applicable to your UDF. Every built-in model's parameter estimates are available to you.
PkProp()
This is the function that will furnish you the scan properties of the peaks for you to use in your own starting estimate formulas. The PkProp(_AREA), PkProp(_CTR) , PkProp(_FWHM), and PkProp(_ASYM50) formulas return the area, the mode (apex) of the peak, the FWHM (full-width at half-maximum), and the Asym50 (half-height asymmetry ratio). Although an a0 area and a1 center are straightforward, you will probably have to find a formula that relates your a2 width and a3 distortion to these or other properties which are computed in real-time and made available to user functions as peaks are placed.
UDF Selection
The dropdown will offer the selection of UDFs 1 through 60. Please note that UDFs may not remain in the position where you install them if lower numbered UDFs are cleared. When you exit the user function procedure, or save a library, the functions will move upward to fill any empty positions. The currently selected UDF will be shown in the little box to the right, as in the number 12 in the screen capture above.
Function Name
This name will be used to represent the function in the function selection list and in the numeric summary. To have these names fit in the Numeric Summary, the length will be limited.
Coefficient Count
This is the number of adjustable parameters in the UDF model. The UDF must contain the number of parameters entered. If, for example, you enter 4 as the coefficient count, the UDF must contain parameters A0,A1,A2,A3 (or a0,a1,a2,a3). The maximum coefficient count is 10.
Entering the UDF
The UDF entry uses a simple ASCII multiline editor. You can use the Cut, Copy, and Paste items to move text about or to paste in the UDF formula if you placed it into the clipboard via another program. All of the functions and constants available within PeakLab can be accessed via the parser help built into the dialog.
Maple and Mathematica
If there isn't too much repetition in the model code, or if you aren't especially concerned with fitting speed, it is probably easiest to copy a final model expression and insert this into the function text field. If you paste expressions from Maple, copy the final expression as text in Maple's language code. For Mathematica, you must copy the final expression as "plain text". For nearly all function expressions, the 8192 character limit should not come into play. The text from Maple should not require any special handling; the PeakLab parser has been extended to handle the Maple text directly. When you paste in text from Mathematica, you will need to specifically use the Paste Mathematica button. This converts Mathematica's syntax to that which is compatible with PeakLab.
Note that certain functions in the Maple and Mathematica function sets may not be available in the PeakLab parser, although every function we found that was applicable to peak modeling should be available. Note also that certain functions in the parser may have two or three different names. One may be compatible with prior versions of the product, another will support Maple's nomenclature, and yet another will support Mathematica's. The functions will be identical and you can use them interchangeably.
Multiple Expressions
Precede the entered or pasted expression with Y= if this the final model. If you enter or paste in components, such as a density and its cumulatives, assign each a variable name. For example, you can assign an expression for the density to Dens= or PDF= or any name you wish. You are limited to nine subexpressions in the overall user function expression. Note that any constants you assign are not counted against this limit. These will be added as global numeric constants to the parser and treated as fixed values. You should thus not assign two different constants to the same name. In a UDF no prefixes are automatically supplied.
Directly Accessing PeakLab's Built-In Functions
UDFs based upon PeakLab equations will execute faster if they use the built-in functions available in the parser. The built-in functions also contain the conditional code ensuring that the functions will return a valid value for all X.
UDFs with Derivative Functions
The first derivative function is DX(FnExprX) and the second derivative function is DX2(FnExprX) where FnExprX is any Fn expression in X. The following UDFs take the first and second derivative of a generalized normal cumulative (the first derivative is thus the generalized normal peak).
Fn1=GenNorm_C(x,a0,a1,a2,a3)
Y=DX(Fn1)
Fn2=GenNorm_C(x,a0,a1,a2,a3)
Y=DX2(Fn2)
UDFs with Integration Functions
The primary integration function is Integrate(FnExpr$,Lower$,Upper$,EpsConverge). It first seeks to achieve the target precision using a successive step Gaussian Quadrature. If this is unsuccessful, a Romberg procedure follows. If the Romberg fails to achieve the desired precision, an Adaptive Quadrature procedure is used. The following example uses the Integrate function to generate the HVL from its derivative.
LOWER=1E-8
FN1=LN($/A2)/A3
FN2=EXP(-0.5*F1*F1)
Y=A0+A1*Integrate(FN2,LOWER,X,1E-6)
Note that the $ symbol is used as the variable of integration. In this example, the second function expression is integrated with $ ranging from a lower limit to X, to a precision of 6 significant figures.
PeakLab also offers an easy way to integrate built in functions. Substitute $, the variable of integration for x in the first argument of the built-in functions. The following function creates a cumulative for the Voigt function:
LOWER=0
FN1=_Voigt($,A0,A1,A2,A3)
Y=Integrate(FN1,LOWER,X,1E-5)
Separate Integration Functions
The Gaussian quadrature, the INTQUAD() function, will use up to 4096 evaluated points for each integration. The adaptive quadrature, the INTADAPT() function. will use to 10,000 evaluated points for each integration. The Romberg algorithm, the INTROMB() function, can use many more, and it is generally the slowest of the three procedures. We recommend the INTQUAD() as the principal algorithm, the INTADAPT() as the first fallback, and the INTROMB() as the final fallback. You must select which integration function is to be used when you create the user-defined model.
Validation
A UDF is extensively validated before it is compiled. If there is a math or parser error, you will be given a clear indication of the error and the cursor will be placed at the location where the validation failed. If you are having trouble with a UDF, you may save the failed UDF to disk. You can then recall it at some future time in an effort to fix what is wrong.
Starting Estimates and Constraints
You must enter starting estimates for the parameters in the model. Since PeakLab UDF's will normally be placed and adjusted within one of the AutoFit procedures, it is important the estimates be sufficient to have the function appear within the X and Y ranges of the data being fitted. Minimum and maximum constraints are optional. If a parameter violates a constraint that you set, a penalty function is added to the chi-square to attempt to bring the parameter back into a valid range.
Entering Formulas for Estimates and Constraints
You may enter formulas for the estimates and lower and upper limits in UDFs. These enable a UDF to be constructed so as to accommodate widely varying X and Y data ranges. Since these formulas are evaluated sequentially, any given formula can reference a previous estimate. For example, A3 can reference A0, A1, or A2, but not A4 and higher. More commonly, a UDF estimate formula will reference an X-Y data table constant which PeakLab computes for each data set. These can consist of the basic X-Y data table constants, such as XMEAN, XSTD, YRANGE, etc. as well additional constants used specifically for UDFs. In the case of multiple data sets, please note that these X-Y data table constants will only apply to the principal data. When such data constants are used in a user function fit, only a single data set should be fitted unless all data sets share very close to the same values.
Since UDFs are often used for fitting single peaks and transitions, PeakLab determines the following data constants:
For peak functions:
XCTR, X at Peak Center
XL50, X at Half-Maxima Left
XR50, X at Half-Maxima Right
XW50, X Width at Half-Maxima (FWHM)
For transition functions:
X50, X at Ymin+Yrange/2
X25, X at Ymin+Yrange/4
X75, X at Ymin+3*Yrange/4
XWTR, X transition width X75-X25
For waveform functions, there are:
XWL, wavelength
XPH, phase for sine
XPH2, phase for sine-squared wave
Note that these constants will not likely have any meaning in data sets containing multiple peaks or transitions.
The constants are specific to the principal data set. Be cautious of using YMAX and XMAX, for example, in user function bounds if there may be different scaled data sets in the file.
Reading a UDF
Use the Read button to read a UDF from disk. The UDF will be read into the current UDF position, replacing any UDF that might currently be in this same position. You may read any UDF into any of the 15 available positions, even if lower numbered positions are empty. This option will read both the ASCII format UDFs and the binary format UDFs containing one or more formulas for estimates or constraints. It is recommended that UDFs be created only within the program.
Clearing a UDF
Use the Clear Current UDF button to immediately free the currently selected UDF from memory and to reset the UDF entry screen.
Saving a UDF
Use the Save button to save a UDF to disk. If the estimates and constraints are numeric, the UDF is saved in an ASCII format with a [UDF] extension. If formulas are present, the UDF is saved in a binary format that requires approximately 4K of disk space. A UDF is always validated before a Save is made. If the validation fails, you are given an option to save the UDF even though it contains an error. You can then recall it at some future time in an effort to repair what is wrong.
Saving a UDF Library
To make it easy to work with up to 15 user functions, any set of installed UDFs can be saved as a user function library. UDF positions are preserved as you enter them and installation of empty UDF positions are permitted. UDF libraries are saved as binary [UDL] files, whether or not estimates contain formulas. In a UDF library, each UDF consumes about 4K, so a full 15 UDF set will require about 60K disk space. To save the current set of UDFs to a library, use the Save UDF Library button. For maximum flexibility, you may wish to save individual UDFs as separate files in addition to having them in libraries. If you choose to keep UDFs only in libraries, you can save out individual UDF components simply by reading the UDL library, selecting the UDF of interest, and then using the individual Save option.
Reading a UDF Library
To read the user functions within a PeakLab UDF library, use the Read UDF Library button. This will install and validate all of the UDFs in the UDL library. If a validation fails, you are notified of such and that specific function is not installed. To extract a UDF from a library, use this option, select the UDF desired, and then use the individual Save option to create the UDF file.
Clearing All UDFs
To clear all of the currently installed UDFs from memory and to clear the current entry screen, use the Clear All UDFs button. You must confirm this option before this the UDFs are freed. Note that it is not necessary to clear the existing UDFs when reading a UDF library since all current UDFs are cleared prior to this read operation.
Parameter Contribution Warning
Even with a successful UDF compilation, you may still get the warning Parameter n Makes Less Than .0001 Fractional Contribution to Equation in X-Range of Data. Adjustment is Recommended. PeakLab determines the minimum and maximum partial derivative for each parameter. This range of partial derivative is multiplied by the current estimate for that parameter and then compared to the overall Y-data range. If a parameter does not make at least a .0001 fractional contribution relative to the Y-data range, this warning is given.
This warning is often a good indicator that the starting estimates are inadequate for placing the UDF within the range of the current data. It may also indicate a poorly designed model that contains a parameter that minimally impacts the equation in the X-range of the data.
If You are Uncertain of Starting Estimates
When working with a new or exploratory model, you may not know what values constitute good starting estimates. In such a case, for each parameter you are uncertain of, simply enter 1.0 for the starting estimate (or some other value for which the UDF is defined). If you achieve a successful compile but get the parameter contribution warning, you will need to adjust the estimate. If you do not get the parameter contribution warning, there is a good chance that the UDF will be placed somewhere within the current range of the data.
Graphical Adjustment of Starting Estimates
The Adjust item is mainly of value for user functions with fixed starting estimates for the parameters. This option opens a graph of the current PeakLab data and the UDF. Do not be surprised if you do not see the UDF at first. If the estimates are too far off, it is possible that no part of the UDF graph will be anywhere within the field of the X-Y data.
If there are formulas for the starting estimates, the goal of graphical adjustment may be to simply set the fixed estimates so that the UDF appears somewhere within the field of the data. The final adjustment of the UDF will occur when it is placed during the Fitting procedure.
You may adjust each parameter with the scrollbar or you may enter the actual value. If you enter an actual value, there will be a brief delay before the screen is updated, allowing you to complete your entry.
For each parameter that has user-specified minimum and maximum constraints, as opposed to the defaults, the scrollbar adjustment will range from this minimum to maximum in 100 steps.
If such constraints are absent, the scrollbar's sensitivity will depend on computed partial derivatives. The partial derivatives are updated with each value entered. If you enter a value for which the partial derivative range is very nearly zero, or if such is true when you begin, the scrollbar adjustment may produce very large jumps. In this case you must enter a value that brings the UDF back into range.
UDF Failures
The two most common causes of UDF failures are having the UDF produce essentially a constant value across the X-range of the data, or to have a parameter set at such a value, that its impact has no impact across the X-range of the data. As you adjust the starting estimates graphically, the UDF is being updated. When you return to the UDF screen, the fixed estimates will reflect those set in the graphical procedure.
Inspecting Partial Derivatives
From the graphical UDF Adjustment screen, you can choose the Derivatives option. This displays a similar set of adjustment controls except instead of seeing a graph, you see a table of partial derivative information.
The % of Y column consists of the % of the Y data range represented by the range of the partial derivative multiplied by the estimate. The last column reports a relative percent based on all non-constant parameters, and these sum to 100%.
The fitting algorithm relies on changes in partial derivatives to guide the revisions in the parameters that occur with each iteration. If a partial derivative is constant, then it should have a value of 1.0, meaning that it is a true constant in the expression. If you see two constants, then at least one of these parameters should be removed. If you find that across wide adjustments of a given parameter there is no significant contribution to the Y-range of the data, whereas the others are making such a contribution, you should seriously question whether this parameter belongs in your model.
Note that you can set the estimate of a given parameter so far out of range that no legitimate partial derivatives are found for any parameter in the X range of the data.


 |