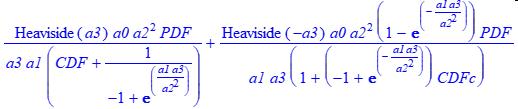
PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
User Function Fitting - Experimental ZDDs
User-Function Fitting of Experimental Zero-Distortion Densities
The universe of potential chromatographic models based on the common chromatographic distortion operator and a zero-distortion density (ZDD) is as vast as an unlimited as the statistical and theoretical probability densities that exist. We feel that we have chosen from the most meaningful densities for chromatographic modeling, but it is an easy matter to extend the existing PeakLab ZDD-based models using user-defined models.
The PeakLab parser includes the following two construction functions to build ZDD-based common distortion chromatographic models:
MHVL(area,ctr,width,distortion,PDF,CDF,CDFc) Generalized Chromatographic Model HVL-a3
MNLC(area,ctr,width,distortion,PDF,CDF,CDFc) Generalized Chromatographic Model NLC-a3
Properly implemented, these used-defined models can be as effective and while not as fast as built-in models which use analytic partial derivatives to guide the fitting, these user defined models can be surprisingly fast.
To create a generalized chromatographic model using your own ZDD, you will need to create both a PDF, a probability density function, the peak model, and either the CDF (or CDF complement), the cumulative distribution function (or reverse cumulative). This means your zero distortion density or peak function will need to be integrable if you wish to have a fast executing closed-form user-defined model.
That mass is conserved is one of the essential foundations of the HVL and NLC models upon which the common chromatographic distortion operator is based. To use this operator at all, you will need to furnish the CDF in either a closed form, or as an integral. For many of the program's peak models, there are closed form cumulatives (which have a _C appended to the peak name), and closed form reverse cumulatives (which have _CR appended to the peak name). The subject of UDF integrals is covered in a separate tutorial. We have limited all ZDDs in PeakLab to models with closed form solutions for the CDFs.
The MHVL Construction Function
The MHVL function uses the HVL a3 parameterization.
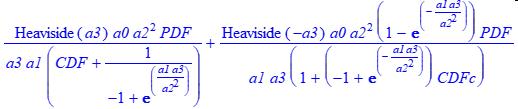
The MNLC Construction Function
The MNLC construction function uses the NLC a3 parameterization.
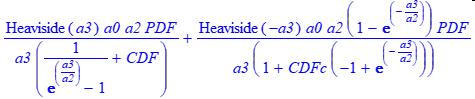
In these models, the first portion of the model is used for +a3 intrinsically tailed peaks, the second for -a3 intrinsically fronted peaks.
In the User Function Fitting - Experimental ZDDs,IRFs (Tutorial), there are UDF examples where the HVL, the GenHVL, and the Gen2HVL are constructed in user functions and fitted.
Managing the Often More Predominant IRF Distortions
Throughout this documentation, we have pointed out the predominance of the IRF (instrument response function) influence on the distortion in chromatographic peaks. It is seldom of value to explore ZDD variations of non-gradient analytical peaks if you don't manage the instrumental/system distortions in some fashion.
When you create any user function, PeakLab automatically creates three additional UDFs with <ge>, <e2> and <e> IRFs. These are added to the internal Fourier fitting of IRF-bearing models. As long as one of these primary IRFs sufficiently models your instrumental/system distortion, you will be able to fit an <irf> variant of your model with no additional effort on your part. You can concentrate on the peak component of the model knowing that you will be able to fit your user-defined peak model with the <ge>, <e2>, or <e> IRF to more accurately characterize these response function parameters. An IRF's parameters will, in general, attempt to fit all delays in the instrument and system that the primary peak model cannot accommodate. There will be small differences in an IRF's parameters even with peak models that offer very strong fits, simply because these different delays in the mass transfer and detection will be apportioned between the primary model and the IRF.
Although you can use the GenHVL<irf> or GenNLC<irf> models to determine your instrument/system response parameters, and then use this IRF in the IRF Deconvolution procedure, this is not recommended if you are researching different densities. The differences between ZDDs are often subtle, and additional sources of error from the IRF complicate the process.
Parametrizations of Built-in Models
Many different parameterizations exist of certain statistical models. For example, the Student's t and Pearson VII models produce identical shapes. The same is true of the Skew Normal and the GMG (half-Gaussian modified Gaussian). Before going to the effort of creating a ZDD in a user-defined model, and possibly going through the labor of deriving the CDF when such isn't readily available, you may wish to use the View Function(X) option. Paste in the ZDD formula from Maple or Mathematica, or enter such directly. Use the Export option to generate a data set which can then be imported, replicated, and fit in a Model Experiment using the different models, usually those in the Statistical family. If you get a perfect fit (typically an F-statistic from 1E+20 to 1E+32, depending on the extent to which the fit captures all sixteen digits of the floating point precision), it is likely your model already exists in the program with a different parameterization. In such a case, you may want to use the built-in model in a UDF where you transform the different variables to replicate the specific parameterization. This will also allow you to take advantage of the program's built-in constraints and error handling, especially on models which are not defined at all x.
User Function Examples
Reproducing the HVL (with built-in functions)
PDF=Gauss(x,1,a1,a2)
CDF=Gauss_C(x,1,a1,a2)
CDFc=Gauss_CR(x,1,a1,a2)
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
Reproducing the HVL (without built-in functions)
PDF=Exp(-((x-a1)^2/(2*a2^2)))/(a2*Sqrt(2*Pi))
CDF=1/2*Erfc(-((x-a1)/(Sqrt(2)*a2)))
CDFc=1/2*Erfc((x-a1)/(Sqrt(2)*a2))
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
Reproducing the GenHVL
PDF=GenNorm[m](x,1,a1,a2,a4)
CDF=GenNorm[m]_C(x,1,a1,a2,a4)
CDFc=GenNorm[m]_CR(x,1,a1,a2,a4)
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
Reproducing the Gen2HVL
PDF=GenError[m](x,1,a1,a2,a4,a5)
CDF=GenError[m]_C(x,1,a1,a2,a4,a5)
CDFc=GenError[m]_CR(x,1,a1,a2,a4,a5)
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
The three primary HVL-based models are constructed using the PDF, CDF, and CDF complements of the Gaussian, Generalized Normal (a1=mean), and Generalized Error (a1=mean) models.
Reproducing the NLC (Fast Approximation)
PDF=Gidx(x,1,a1,a2)
CDF=Gidx_C(x,1,a1,a2)
CDFc=Gidx_CR(x,1,a1,a2)
Y=MNLC(a0,a1,a2,a3,PDF,CDF,CDFc)
Reproducing the NLC (Exact)
PDF=Giddings(x,1,a1,a2)
CDFc=TFn(a1/a2,x/a2)
CDF=1-CDFc
Y=MNLC(a0,a1,a2,a3,PDF,CDF,CDFc)
Reproducing the NLC (using Generalized Normal)
v2=sqrt(2*a1*a2)
v4=sqrt(2)*sqrt(a2/a1)*0.5
PDF=GenNorm[m](x,1,a1,v2,v4)
CDF=GenNorm[m]_C(x,1,a1,v2,v4)
CDFc=GenNorm[m]_CR(x,1,a1,v2,v4)
Y=MNLC(a0,a1,a2,a3,PDF,CDF,CDFc)
Reproducing the GenNLC
v2=sqrt(2*a1*a2)
v3=a3/2
v4=sqrt(2)*sqrt(a2/a1)*a4
PDF=GenNorm[m](x,1,a1,v2,v4)
CDF=GenNorm[m]_C(x,1,a1,v2,v4)
CDFc=GenNorm[m]_CR(x,1,a1,v2,v4)
Y=MNLC(a0,a1,a2,v3,PDF,CDF,CDFc)
Reproducing the Gen2NLC
v2=sqrt(2*a1*a2)
v3=a3/2
v5=sqrt(2)*sqrt(a2/a1)*a5
PDF=GenError[m](x,1,a1,v2,a4,v5)
CDF=GenError[m]_C(x,1,a1,v2,a4,v5)
CDFc=GenError[m]_CR(x,1,a1,v2,a4,v5)
Y=MNLC(a0,a1,a2,v3,PDF,CDF,CDFc)
Using the equivalence relationships which convert between the diffusion-based HVL and the kinetics-based NLC, the Generalized NLC models are built using the same PDF, CDF, and CDF complements of the Generalized Normal (a1=mean), and Generalized Error (a1=mean) models. The adjustment in a3 for the Generalized NLC models is done so that the GenHVL and GenNLC share an identical a3 distortion value.
Creating an Entirely New Chromatographic Model
It is recommended than any ZDD you use be capable of reducing to a Gaussian. At such parameter values, the chromatographic model will reduce to the HVL. For this example we will use the GEaMG model built into the program. Like the EMG, the GEaMG is the convolution of a Gaussian with a delay component. Here the delay is an area weighted sum of an exponential and half-Gaussian instead of a simple exponential. There are two widths, an SD for the half-Gaussian and a time constant for the exponential as well as an area fraction for the first component. As these two widths approach zero, the model becomes a Gaussian. This is the user-function example we used in the Common Distortion Model topic.
PDF=GEaMG(x,1,a1,a2,a4,a5,a6)
CDF=GEaMG_C(x,1,a1,a2,a4,a5,a6)
CDFc=GEaMG_CR(x,1,a1,a2,a4,a5,a6)
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
The PDF must have unit area, and the CDF and CDFc must have unit amplitude. The a1 center, and a2 widths will exist in the PDF, CDF, and CDFc as well as in the MHVL function. The a3 parameter, the chromatographic distortion, will be specific to the MHVL function. Any parameter sequence beyond a2 must start at a4 onward. The a3 parameter must not appear in the PDF, CDF, and CDFc expressions.
Using Mathematica or Maple for new ZDD Models
For more complex expressions where there is no built-in PeakLab model, we recommend building the PDF, CDF, and CDFc in Maple or Mathematica and graphing/testing it there prior to building a PeakLab user-defined peak. The guidance for importing Maple or Mathematica expressions into PeakLab is given in the user functions topic.


 |