PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
ZDD Concepts
In order to understand the Zero-Distortion Density (ZDD) enhancements to PeakLab's fitting, you must first understand the HVL and NLC chromatographic models.
The HVL produces a theoretical diffusion width, generally seen as applicable to GC, and the NLC produces a kinetic time constant, generally seen as applicable to LC when slow kinetics of adsorption and desorption are present, or where mass-transfer can be modeled by such kinetics.
The HVL Chromatographic Model
The Haarhoff-Van der Linde (“HVL”) gas chromatography model is defined as follows:
(1)
If we look at the HVL as a statistical model absent its theoretical derivations, and in a form where the area is an adjustable parameter, we have a four parameter function. In this form, a0 is the peak area, a1 is the center or location value, a2 is the peak width or scale parameter, and a3 is the shape parameter, positive for a right-skewed asymmetry, negative for left-skewed.
The NLC Chromatographic Model
The Wade-Thomas non-linear liquid chromatography model (“NLC”) is defined as follows:
(2)
Where TFn is a Bessel function integral:
(3)
When the area is an adjustable parameter, the NLC is also a four parameter function. As with the HVL, a0 is the peak area and a1 is the center or location value, and a3 is the distortion parameter, positive for the right-skewed asymmetry of a tailed peak and negative for the left-skewed asymmetry of a fronted peak.
This NLC parameterization uses a time constant instead of a rate constant for the a2 kinetic parameter given in the original publication of the Wade-Thomas NLC model. As such, the NLC a2 is similar to the HVL a2, a scale parameter that increases with the peak width.
The Generalized HVL Template
In order to generalize the HVL model, we use the Gaussian or normal probability density function (PDF), normalized to unit area:
(4)
We also use the Gaussian or normal cumulative distribution function (CDF):
(5)
We also take note of the complement of the CDF, the reverse cumulative of the normal density, even though it is not used in the HVL:
(6)
We can now rewrite the HVL as a generalized template that accepts any zero-distortion density:
(7)
To regenerate the HVL, Density is replaced with (4), the normal PDF, and Cumulative with (5), the normal CDF. Note that any replacement is always done with a matched PDF-CDF (Density-Cumulative) pair.
The Generalized NLC Template
To create a generalized NLC model template, we use the Giddings density:
(8)
Here we take note of the Giddings cumulative, although it is not used in the NLC:
(9)
We also use the Giddings reverse cumulative:
(10)
The NLC density generalized template can then be expressed as follows:
(11)
Just as with the HVL template, we can create any number of NLC generalized models by inserting a matched density-cumulative pair other than the Giddings for the zero-distortion assumption.
To regenerate the NLC, Density is replaced with (8), the Giddings PDF, and RevCumulative with (10), the Giddings CDF complement.
The Common Distortion Model
Although one of these templates uses a CDF and the other a reverse CDF, the distortion produced by the models can produce identical shapes for a given density-cumulative pair. One can substitute the Gaussian density in the NLC template to generate the HVL shape, and the Giddings density can be inserted into the HVL template to produce the NLC shape.
Most of PeakLab's best chromatography peak models are based on this common distortion model. These two templates are used interchangeably within PeakLab, where possible, to construct models with a maximum dynamic range of a3 chromatographic distortions.
Note that the a1 associated with the first moment, and the a2 associated with the second moment also appear in the templates, and while the a1 center values are comparable, the a2's consist of immensely different representations of the peak widths, one a Gaussian diffusion width, the other a kinetic time constant associated with adsorption-desorption.
Because the generalized HVL (GenHVL) and generalized NLC (GenNLC) models can produce exactly the same shapes, it is possible to convert a fitted a2 so that it represents either an HVL diffusion width or an NLC kinetic time constant. This is what is given in the Equivalent Parameters section of the Numeric Summary of a fit.
Higher Moments Adjustments
The major drawback of the basic HVL and NLC models is that the higher moments are fixed by the Gaussian and Giddings zero-distortion assumptions.
If you have long used both the HVL and NLC models in fitting chromatographic peaks, you were probably struck by their similarities. Part of this can be attributed to the similarity between the Gaussian and Giddings zero-distortion densities:
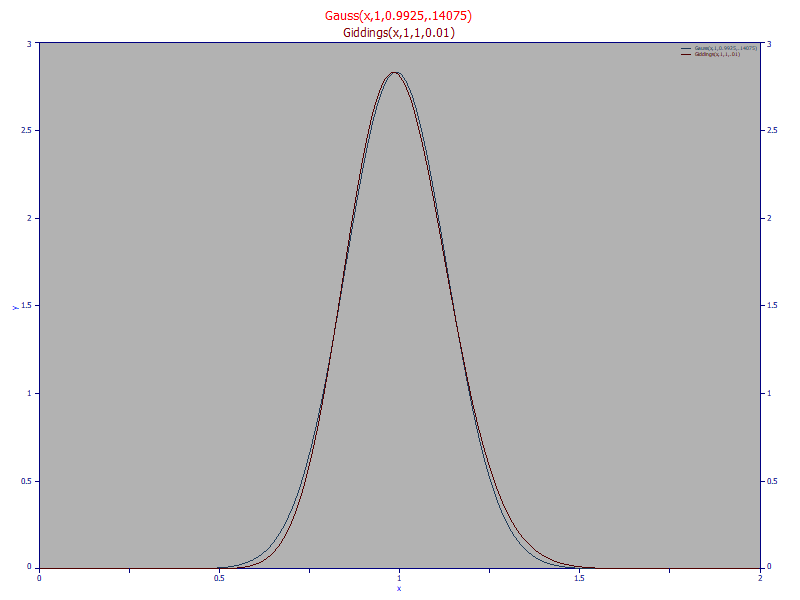
The Giddings density, the red curve, is a slightly right-asymmetric peak as compared to the symmetric Gaussian, the blue curve. This symmetry explains why the HVL produces mirrored shapes about a1 with negative distortions, whereas the NLC produces different tailed and fronted shapes with the same magnitude of the a3 distortion parameter.
The HVL and NLC generalized templates allow for any density-cumulative pair to be used. In order to deal with the non-idealities in the actual chromatographic separation, we assume the ZDD is neither Gaussian (HVL) or Giddings (NLC) but a more complex density that allows for the third moment, the skew to be broadly adjustable. This is the premise of most of the ZDD models in PeakLab.
ZDD Interpretations
Since the ZDD accounts deviations from the theoretically expected third and/or fourth moments, the interpretation of the ZDD adjustments is a matter of choice:
1. The deviations from a Gaussian ZDD can be deemed non-idealities in the chromatographic separation, and the a2 width can be taken as an absolute diffusion or Gaussian broadening.
2. The deviations from a Giddings ZDD can be deemed non-idealities in the chromatographic separation, and here the a2 width can be taken as a true kinetic term, possibly one where lumped contributions to a2 are removed in deconvolving the Giddings ZDD.
In both of these cases, the terms associated with the skew and kurtosis of the ZDD can be dismissed, and seen merely as tools for deconvolving a truer a2 parameter. Also, in both cases, the a3 will be identical, and for analytical peaks, generally linear with concentration.
It is also possible to use the higher moment information:
3. The moments of the actual ZDD can be used as proxies for the absolute moments of the peak at infinite dilution. The extent to which the ZDD's higher moments stray from expectation may yield useful information in designing preps.
4. Since modeling the higher moments and deconvolving or fitting the IRF can result in close to zero-error fits, one can also choose to use the actual moments of the fitted peak as the key parameters of interest. For a given standard, those moments should be constant across time, and any deviation may be a good indicator of column degradation.
ZDD Considerations
1. There is one primary ZDD in the program, used by both the GenHVL and GenNLC basic family of models (where no [ZDD] suffix is added). The 'Gen' ZDD models are 'once-generalized', using an asymmetric generalized Normal which allows the skew or third moment to vary across a wide range. For most non-gradient analytical peaks, the default GenHVL and GenNLC models will suffice and in our experience, no other models in the program are likely to produce better fits. For analytical LC, it is possible the GenNLC is the only model that will ever be needed.
2. There is a second ZDD of major importance, especially in gradient HPLC and fitting overloaded peaks. This is the generalized Error model used in the Gen2HVL and Gen2NLC family of models (similarly, no [ZDD] suffix is used). The 'Gen2' ZDD models are 'twice generalized', containing two generalizations, one that adjusts the skew or third moment, and another that adjusts the kurtosis or fourth moment. The latter is sometimes described as adjusting the 'fatness' of the tails of a density. This fourth moment kurtosis adjustment will definitely be needed for gradient peaks since it will reflect the compression of the peak decay arising from the gradient. Such fits may be of value in detecting any anomaly in the expected gradient. If the gradients are accurate, the peaks will reflect a consistent compression in this fourth moment parameter. You should expect the a4 power term to increase from the non-gradient 2.0 tail decay to values ranging from 2.1-2.3. This may seem small, but in statistical terms, it is a huge compression in the decay, especially given that it also hides the right-shifted instrumental distortions.
3. For all other ZDDs, a nomenclature similar to GenHVL[ZDD] is used. The ZDD-IRF nomenclature is separate and unambiguous. A model designated as GenHVL[Y]<ge> consists of the 'Y' generalized error ZDD convolved with a 'ge' half-Gaussian/exponential IRF.
4. The GenHVL's a2 consists of a Gaussian diffusion width and its a4 consists of a statistical asymmetry. The GenNLC's a2 consists of a Giddings kinetic time constant and its a4 is normalized to 0 for a Gaussian ZDD and 0.5 for a Giddings ZDD. Both the GenHVL and GenNLC can produce identical peak shapes when individually fitted and the a4 parameters are not shared across all peaks, and both can model pure HVL and NLC shapes as well as those arising from a variable skew.
5. For the GenHVL and GenNLC models, the only differences are in a2's actual value and interpretation and in the a4 skew's normalization. For the GenHVL, the a4 is an absolute statistical asymmetry. For the GenNLC, the a4 is indexed to the Giddings asymmetry where an a4 of 0.5 always produces the NLC's Giddings ZDD. For LC, and where a4 is shared across all peaks, the GenNLC allows all peaks to be pure NLCs with a constant 0.5 a4. Such will not be possible across multiple peaks of the GenHVL with a common shared a4. An a4 of 0 for the GenNLC is a Gaussian ZDD (an HVL). An a4 higher than 0.5 has a higher skew than would be predicted by the NLC theory. This GenNLC is capable of approximating the pure Bessel-integral NLC to at least six significant digits, appreciably better than any current chromatographic instrument, and its speed is comparable to the GenHVL. There is no longer any form of computational penalty for using an NLC-type model.
6. The GenHVL uses the statistical asymmetry a4 parameter to control the skew, a positive value for adding a right skew to the ZDD, a negative value for adding a left skew.
7. PeakLab offers a [Z] suffix ZDD, for both the GenHVL and GenNLC, which consists of this same generalized normal with a parameterization where the a1 is the mean of the underlying normal in the ZDD instead of the mean of the asymmetric ZDD density. This represents a full statistical deconvolution to a fitted Gaussian width. Similarly, PeakLab offers a [Y] suffix ZDD. This ZDD is identical to the ZDD used in the Gen2HVL and Gen2NLC models, except the generalized error model is also used with this parameterization where the a1 is the mean of the underlying normal in the ZDD.
8. A number of ZDDs were retained in the product from the research carried out to create this product. If any of these demonstrated even modest benefits in modeling the deviations from the Gaussian or Giddings theoretical densities, these were retained in the product. Apart from the primary GenHVL and GenNLC models, Gen2HVL and Gen2NLC models, and the similar [Z] and [Y] ZDD models, all other ZDD's should be deemed experimental.
9. As the complexity of a ZDD increases to include the modeling of one or two higher moments, a strong S/N will be needed in the data. Peaks with significant noise will probably have to be modeled with basic NLC or HVL models, perhaps even EMG or simple Gaussian models. The parameter statistics in a fit will clearly show where models are overspecified.
Built-In ZDD
The following models are used in the product to extend the modeling power of the Gaussian ZDD (HVL) and the Giddings ZDD (NLC):
Default Asymmetric Error Model
[G] Half-Gaussian Modified Gaussian (GMG)
[E] Exponentially-Modified Gaussian (EMG)
[T] Generalized Student's t (Asymmetric)
[Yp] Generalized Error Model (High Overload Shapes Only)
[YpE] Generalized
Laplace (High Overload Shapes Only)
[Yp2]
Generalized Error 2-Width (Overload Shapes)
[Yp2E] Generalized Laplace 2-Width (High Overload Shapes Only)
[K] Generalized Two-Sided Kinetic Orders 0,1 (High Overload Shapes Only)


 |