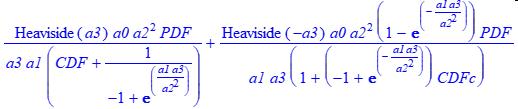
PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
User Function Fitting - Experimental ZDDs,IRFs (Tutorial)
Although user-defined peaks can be constructed for any model you wish to fit, in this tutorial we will focus on the specific instances where user-defined models are most likely to prove useful in extending the technologies within PeakLab.
Alternate ZDD - Zero Distortion Densities
In fitting a chromatographic model to close to zero error, we found it necessary to employ the common chromatographic distortion with higher moment statistical generalizations of a zero-distortion density (ZDD). For practical reasons, we limited our implementation of more generalized ZDDs to closed form models. We did not explore ZDDs which could only be expressed in integral form. In most cases, such integrals described convolution components which could be treated separately as IRFs in the program's fitting of convolution integrals in the Fourier domain. We also omitted non-closed form ZDDs for another reason: we had achieved near perfect fits without them. Not surprising, the best ZDDs were those which fully generalized the HVL and NLC models by being fully capable of rendering both the Gaussian and Giddings densities.
Before authoring an experimental ZDD, please take a close look at the different ZDDs built-in to the program. These are also listed in the Generalized HVL Models and Generalized NLC Models help topics. The ZDD you are looking for may already be in the program or the density you are looking at may be a specialization of one of these ZDDs which adjust the third and/or fourth moments of the density. Before creating a unique ZDD user model, you may wish to use the Search for Optimum Model feature with a data set containing a single peak, or create a Model Experiment with replicates of a data set of one or more test peaks, in order to explore the built-in models in a single step. The Search for Optimal Model can fit hundreds of potential models in a single unattended step, and any installed user-functions are automatically included, but this exploratory fitting is limited to data containing a single peak.
With few exceptions, a complete chromatographic model is a convolution integral of a peak function (usually this common chromatographic distortion applied to a ZDD) and an IRF, an instrument response function. In general, that convolution integral will not have a closed form solution. An IRF is normally just a decay/delay component (or a sum of two decay/delay components) which represent flow system, detector, and other system distortions unrelated to the pure chromatographic separation. PeakLab contains a number of built-in IRFs, both single and two-component, and with a Fourier domain processing these models will fit far faster than any user-defined function which contains a convolution integral for the IRF.
To make the IRF aspect of modeling a non-issue, and to make the exploration of different ZDDs as simple and as effective as possible, all user defined functions are automatically created with <e>, <g>, <ge>, <e2>, and <v> IRFs. Unless you are specifically researching your own IRF model, you only need to select one of these user function variants at the time of fitting. No specific IRF need be included in your model.
In the first part of the user function fitting tutorial, we will regenerate the main chromatographic models in the program. We will also fit an experimental model.
User-Function Fitting of Experimental Zero-Distortion Densities
The universe of potential chromatographic models based on the common chromatographic distortion operator and a zero-distortion density (ZDD) is as vast and unlimited as the statistical and theoretical probability densities that exist. To make the construction of these models as simple as possible, the PeakLab parser includes the following two construction functions to build ZDD-based common distortion chromatographic models:
MHVL(area,ctr,width,distortion,PDF,CDF,CDFc) Generalized Chromatographic Model using
HVL-a3
MNLC(area,ctr,width,distortion,PDF,CDF,CDFc) Generalized Chromatographic Model using
NLC-a3
Properly implemented, these user-defined models can be as effective, and while not as efficient as built-in models which use analytic partial derivatives to guide the fitting, these user defined models can be surprisingly fast.
To create a generalized chromatographic model using your own ZDD, you will need to create a PDF, a probability density function, the peak model, the CDF, the cumulative distribution function, and the CDFc, CDF complement or reverse cumulative. This means your zero distortion density or peak function will need to be integrable if you wish to have a fast executing closed-form user-defined model.
That mass is conserved is one of the essential foundations of the HVL and NLC models upon which the common chromatographic distortion operator is based. To use this operator, you will need to furnish the CDF in either a closed form, or as an integral. For many of the program's peak models, there are closed form cumulatives (which have a _C appended to the peak name), and closed form reverse cumulatives (which have _CR appended to the peak name). We have limited all ZDDs in PeakLab to models with closed form solutions for not only the PDFs, but the CDFs as well.
The MHVL Construction Function
The MHVL function uses the HVL a3 parameterization.
In these models, the first portion of the model is used for +a3 intrinsically tailed peaks, the second for -a3 intrinsically fronted peaks.
We will now import a PeakLab data file containing a user-function library which makes use of these construction models to regenerate the HVL, GenHVL, and Gen2HVL models.
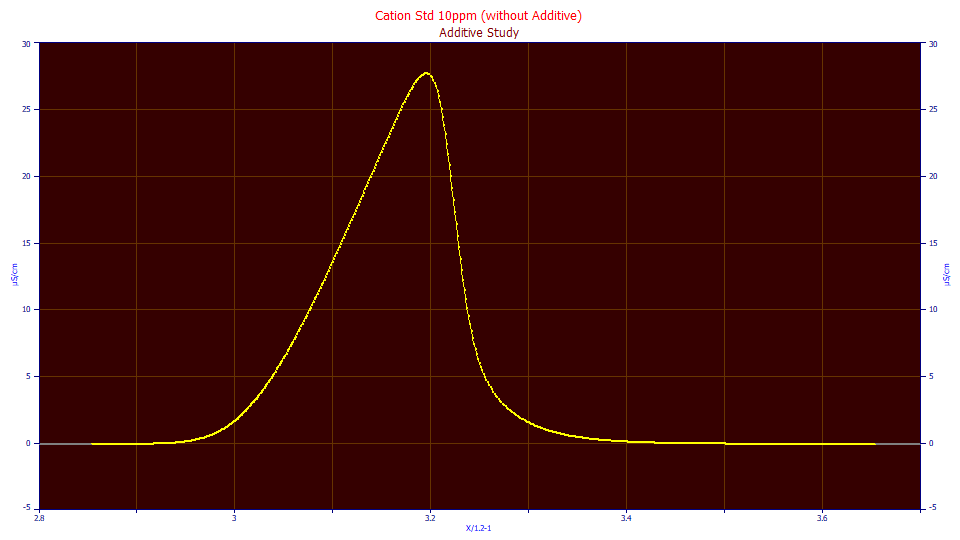
Sample Data for an ZDD/IRF Experiment
![]() From the File menu, select Open... (or use the main toolbar Open button) and the
choose the file UDF_ZDD_Tutorial.pfd from the program's installed default data directory (\PeakLab\Data).
From the File menu, select Open... (or use the main toolbar Open button) and the
choose the file UDF_ZDD_Tutorial.pfd from the program's installed default data directory (\PeakLab\Data).
![]() From the Peaks menu, select User Functions... (or use the main toolbar User Function
button). Any user functions present when a file is saved are included in the PeakLab pfd data file.
From the Peaks menu, select User Functions... (or use the main toolbar User Function
button). Any user functions present when a file is saved are included in the PeakLab pfd data file.
We will initially use the first three user defined functions in the file.
Replicating the HVL Model with a User Function
The first user function replicates the HVL using the MHVL construction function and the PDF, CDF, and CDF complement of the normal density.
PDF=Exp(-((x-a1)^2/(2*a2^2)))/(a2*Sqrt(2*Pi))
CDF=1/2*Erfc(-((x-a1)/(Sqrt(2)*a2)))
CDFc=1/2*Erfc((x-a1)/(Sqrt(2)*a2))
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
In this case, we have used the actual formulas for the Gaussian PDF, CDF, and CDFc. Note that the a1 center value and a2 width value, of the resultant peak, appear in both the MHVL construction function and in the PDF, CDF, and CDFc formulas.
Note also that the starting estimates are not fixed values, but formulas that use the program's automatic placement of HVL peaks:
PkFnParm(HVL,0)
PkFnParm(HVL,1)
PkFnParm(HVL,2)
PkFnParm(HVL,3)
We use the PkFnParm function to 'borrow' the program's starting estimate algorithm for the HVL a0 area, a1 center, a2 width, and a3 distortion values.
We gave this user function the name HVL. There will be a user function number appended so there is no risk of conflict with the built-in HVL function. We also specify the count of parameters as 4, and we have adjusted the bounds of the a3 distortion to vary between -1 and 1.
Replicating the GenHVL Model with a User Function
Select 2 in the Selected UDF dropdown. The current content of the first UDF is compiled and validated. If the validation had failed, you would have remained in the first UDF to make the necessary corrections.
PDF=GenNorm[m](x,1,a1,a2,a4)
CDF=GenNorm[m]_C(x,1,a1,a2,a4)
CDFc=GenNorm[m]_CR(x,1,a1,a2,a4)
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
The construction process for the GenHVL is similar, but there are two important differences to note. Here we use the built in GenNorm[m], GenNorm[m]_C, and GenNorm[m]_CR generalized normal models for the PDF, CDF, and CDFc. We also skip the a3 intrinsic chromatographic distortion parameter in the PDF, CDF, or CDFc parameter sequence. The a3 chromatographic distortion is used only in the MHVL construction function. Any higher moment related parameters must thus begin with a4. In this second UDF we use the PkFnParm function for starting estimates using the GenHVL model, again exactly replicating the starting estimates that the program uses for the GenHVL model for each of the parameters. Here the third moment a4 statistical asymmetry is also bounded between -1 and 1. The function now has a parameter count 5.
Replicating the Gen2HVL Model with a User Function
Select 3 in the Selected UDF dropdown.
PDF=GenError[m](x,1,a1,a2,a4,a5)
CDF=GenError[m]_C(x,1,a1,a2,a4,a5)
CDFc=GenError[m]_CR(x,1,a1,a2,a4,a5)
Y=MHVL(a0,a1,a2,a3,PDF,CDF,CDFc)
The construction process for the Gen2HVL is identical to the once-generalized GenHVL UDF, except we use the GenError[m] set of built in functions instead of the GenNorm[m] functions. Note that the additional higher moment parameters are a4,a5 and that we now have a 6 parameter UDF, and PkFnParm starting estimates borrowed from the program's Gen2HVL model. In the built-in Gen2HVL and Gen2NLC models, a4 is the fourth moment power term and a5 is the third moment asymmetry.
Click OK to return to the main menu.
Testing the User-Defined Functions in a Model Experiment
Right click the main graph and select the Replicate this Data Set a Specified Number of Times from the popup menu. Enter 5 for the count of replicates and click OK. You should now have six identical data sets in the main window.
![]() Click the Local
Maxima Peaks button in the main window (or you can use the Fit Local Maxima Peaks... in
the Peaks menu or main toolbar).
Click the Local
Maxima Peaks button in the main window (or you can use the Fit Local Maxima Peaks... in
the Peaks menu or main toolbar).
![]() Click the AI Expert option to have the program select an optimum smoothing level. It will be small
for this high S/N set of just over 500 points, a window of 7 total points.
Click the AI Expert option to have the program select an optimum smoothing level. It will be small
for this high S/N set of just over 500 points, a window of 7 total points.
The placement options are the defaults:
Peak Type
Select Chromatography
in the first dropdown
Select HVL as the model in the second dropdown
Scan
Set the Amp %
threshold to 1.5 %
Leave Use Baseline Segments unchecked
Be sure Use IRF,ZDD is checked
Vary
Leave width a2
and shape a3 checked, all other unchecked
Click the IRF button. If you made modifications to the IRF defaults prior to this tutorial for your own fitting, please use the Save button to save your IRF values before resetting the defaults. Click the Defaults button. These are based on retention scale averages of modern IC data for each of the IRFs built into the program. Click OK.
Click the Model Experiment button. In a model experiment, multiple models are fit to identical replicates of data allowing you to rapidly compare the different models in a single step.
Select the following models from near the top of the list:
HVL<ge>
GenHVL<ge>
Gen2HVL<ge>
Add the following from near the bottom of the list:
HVL<ge>-udf1
GenHVL<ge>-udf2
Gen2HVL<ge>-udf3
The dialog will show Models (6 selected). Click OK. There are now six different placements based on the six models in the model experiment. Three are the built-in models; three are user functions.
At this point, you must not change any of the items in the dialog. If you do so, a new scan will be made using the selected model and the model experiment will be lost.
Click the Peak Fit button in the lower left of the dialog to open the fit strategy dialog. Select the Fit with Reduced Data Prefit, 2 Pass, Lock Shared Parameters on Pass 1. Be sure the Fit using Sequential Constraints box checked. Click OK.
The fits should only require a second or two. When the fitting is finished, note the penalty for using the user function. Although the fitting of all six models begins simultaneously, the time reported for each is when that specific fit completed. You should see the same iteration count for both the built-in model and the user function, and the user function fitting times should be about 20% slower.
Click Review Fit. As you will note, we have exactly replicated the three primary HVL-based functions in the program using the generalized HVL construction function and the three different densities. The once and twice generalized models, with both ZDD and IRF, fit to less than 2 ppm unaccounted variance. The HVL<ge> model, with an IRF but no higher moment ZDD adjustment, fits to 79 ppm error.
Select IRF Deconv. in the second dropdown. Note that the program is aware of the <ge> IRF and removes such from the user functions just as it removes such from the built-in functions.
Select Partial Deconv. in the third dropdown. Note that you see a Gaussian in green for the HVL<ge> in the first data set, an HVL in green for the GenHVL<ge> and Gen2HVL<ge> in the second and third data sets, and only a repeat of the IRF-removed data in the user function data sets. The ZDD partial and full deconvolution rendering is only available for built-in functions.
Double click the second graph.
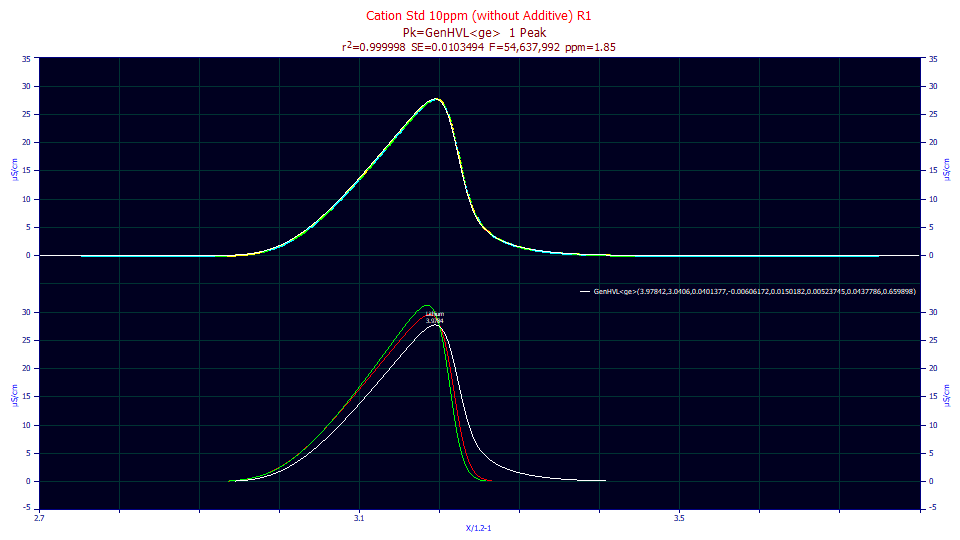
Note the difference between the pure HVL in green and the eluted peak minus the IRF, the GenHVL, in red. We will focus to this difference in the next part of this tutorial.
Double click the graph to restore all plots. Click OK to exit the Review. Change the name to Model Experiment and click OK. Click OK to acknowledge the change to the data file. Click OK to return to the main menu.
Creating a Custom Peak Function
We will now create a new user-defined peak. Let us say we wish to fit an HVL peak which assumes not a delta injection, but a rectangular input. In this case, we don't want to create a different IRF. We simply want a custom HVL which assumes a fixed width for the injection instead of a delta input. We want to know if modifying the core peak in this way improves fitting. Specifically, we wish to know if the difference between the 79 ppm error in the HVL<ge> fit above and the 2 ppm error in the GenHVL<ge> was due to the GenHVL ZDD accommodating the distortion associated with an injection width.
For such exploration of new peak models, we recommend doing the symbolic math in Mathematica or Maple, testing and plotting the model there, and transferring it to PeakLab once you are certain everything is working.
This example is one of the rare instances where it is actually possible to create a closed form solution using the HVL as one component of a convolution integral:
Creating a Custom Peak Function from a Mathematica or Maple Formula
Most of the real-valued functions in Maple and Mathematica which are useful in peak density models are available in the PeakLab's user functions. The program's parser has been extended so that most Maple formulas can be pasted directly into user function expressions without modification.
For Mathematica, a special import is required. If we highlight the above formula in Mathematica, and use the Copy As | Plain Text right click option within Mathematica, the following string is copied:
(a0 a2^2 (Log[(2+(-1+E^((a1 a3)/a2^2)) Erfc[(a1-x)/(Sqrt[2] a2)])/(2+(-1+E^((a1 a3)/a2^2)) Erfc[(a1+a4-x)/(Sqrt[2] a2)])]))/(a1 a3 a4)
To have this work within a user function, you must place the cursor in the Function area of the user function dialog, and click the Paste Mathematica button. The expression is converted as follows:
(a0*a2^2*(Log((2+(-1+E^((a1*a3)/a2^2))*Erfc((a1-x)/(Sqrt(2)*a2)))/(2+(-1+E^((a1*a3)/a2^2))*Erfc((a1+a4-x)/(Sqrt(2)*a2))))))/(a1*a3*a4)
For all user functions, you must place Y= at the beginning of the final peak model expression.
![]() From the Peaks menu, select User Functions... (or use the main toolbar User Function
button). Change the Selected UDF to 4.
From the Peaks menu, select User Functions... (or use the main toolbar User Function
button). Change the Selected UDF to 4.
This user function, assigned the name "RHVL", contains this rectangular injection width variant of the HVL from the formula above.
We have set a starting estimate of 0.1 minute for the injection width as it appears at the column.
Click OK to exit the UDF procedure.
Visualizing User Functions
Although it is usually best to graph your functions in Maple or Mathematica, we will test the UDF before fitting within PeakLab in order to visualize the influence of the rectangular input pulse on the HVL shape.
Select View Function(X) from the Data menu. At the bottom of the dialog, enter 1 in the A0 field, 4 in the A1 field, .1 in the A2 field, and .05 in the A3 field. Change the Min X to 3 and the Max X to 5.
For the Y[1] through Y[5] functions, paste in the following:
UDF5P(4,x,a0,a1,a2,a3,1e-5)
UDF5P(4,x,a0,a1,a2,a3,.025)
UDF5P(4,x,a0,a1,a2,a3,.05)
UDF5P(4,x,a0,a1,a2,a3,.1)
UDF5P(4,x,a0,a1,a2,a3,.2)
The UDF5P parser function processes 5 UDF parameters (a0-a4). The first argument specifies UDF 4. Here we vary the a4 injection width from effectively zero to 0.20 min (12 sec). Click OK.
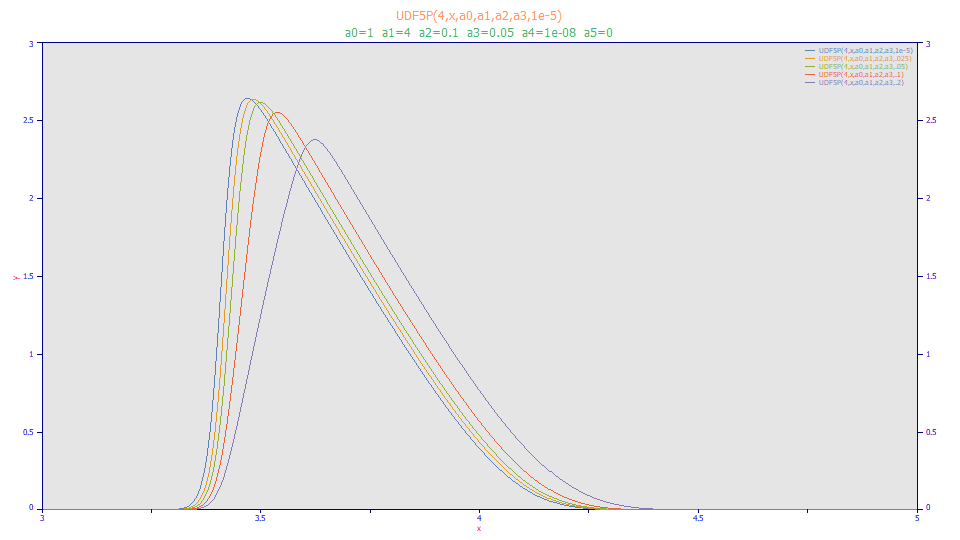
Here we see that the rectangular injection pulse assumption can indeed change the peak shape, a broadening and attenuation, but there is mostly a locational shift until the rectangular pulse is of a 3 second duration or greater. For this model to be valid, one would have to assume the injection width broadens that greatly by the time the pulse reaches the column.
Click OK to close the View Function procedure.
Click Unselect All from the main window. Only the principal data set remains selected. All others now have a grayed background. Click the second graph to reselect it. Its background will again turn blue. Only selected sets are passed to the fitting procedure.
![]() Click the Local
Maxima Peaks button in the main window (or you can use the Fit Local Maxima Peaks... in
the Peaks menu or main toolbar).
Click the Local
Maxima Peaks button in the main window (or you can use the Fit Local Maxima Peaks... in
the Peaks menu or main toolbar).
The previous settings are remembered.
Click the IRF button. Check the Lock button next to the <ge> IRF's g area frac parameter. We don't want any part of an injection effect to be masked by a change in proportion of the two components of the IRF. Click OK.
Click the Model Experiment button. Select the following models from near the bottom of the list: HVL<ge>-udf1 and RHVL<ge>-udf4. The dialog will show Models (2 selected). Click OK.
Click the Peak Fit button in the lower left of the dialog to open the fit strategy dialog. Select the Fit with Reduced Data Prefit, 2 Pass, Lock Shared Parameters on Pass 1. Be sure the Fit using Sequential Constraints box checked. Click OK.
Click Review Fit when the fitting is complete.
Click Numeric and from the Options menu of the Numeric Summary, choose Select only Fitted Parameters and then choose Parameter Statistics:
"Cation Std 10ppm (without Additive)"
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99990198 0.99990115 0.07520463 1,448,525 98.0211310
Peak Type a0 a1 a2 a3 a4 a5 a6
1 HVL-udf1<ge> 3.98372974 3.03380864 0.04160484 -0.0066186 0.01004954 0.04210364 0.61132500
Parameter Statistics
Peak 1 HVL-udf1<ge>
Parameter Value Std Error t-value 95% Conf Lo 95% Conf Hi
a0 3.98372974 0.00187479 2124.89645 3.98004895 3.98741053
a1 3.03380864 0.00072558 4181.22452 3.03238411 3.03523318
a2 0.04160484 0.00015270 272.464774 0.04130505 0.04190463
a3 -0.0066186 1.607e-5 -411.85574 -0.0066501 -0.0065870
a4 0.01004954 0.00071531 14.0492023 0.00864516 0.01145392
a5 0.04210364 0.00036277 116.060420 0.04139140 0.04281587
a6 0.61132500
"Cation Std 10ppm (without Additive) R1"
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99990223 0.99990126 0.07516174 1,208,482 97.7714683
Peak Type a0 a1 a2 a3 a4 a5 a6 a7
1 RHVL-udf4<ge> 3.98338349 3.02451829 0.04066720 -0.0067029 0.01540540 0.00991304 0.04216374 0.61132500
Parameter Statistics
Peak 1 RHVL-udf4<ge>
Parameter Value Std Error t-value 95% Conf Lo 95% Conf Hi
a0 3.98338349 0.00194536 2047.63817 3.97956414 3.98720283
a1 3.02451829 0.00248287 1218.15202 3.01964362 3.02939295
a2 0.04066720 0.00042890 94.8169750 0.03982513 0.04150927
a3 -0.0067029 3.5607e-5 -188.24909 -0.0067728 -0.0066330
a4 0.01540540 0.00423865 3.63450684 0.00708359 0.02372721
a5 0.00991304 0.00102510 9.67028013 0.00790044 0.01192565
a6 0.04216374 0.00045950 91.7599651 0.04126159 0.04306588
a7 0.61132500
We are nowhere close to the near zero error we observed with the GenHVL<ge> model in the previous fits. In fact, the error is virtually unchanged by introducing a rectangular injection pulse into the core HVL model. The F-statistic actually diminishes, suggesting the additional parameter was detrimental rather than beneficial with respect to modeling power. Although all parameters fit as significant, the injection width fits to the lowest significance of all of the parameters (see the 3.63 t-value). At an a4 of .0154, the fitted injection width is less than a second.
Click OK in the main Review window to close the Review. You will be asked to save the fits in the data file. Enter RHVL Study for the name and click OK and again click OK to confirm that the fits were saved. You will be returned to the placement screen. Click OK to return to the main window.
The Delta Input Assumption
Have we actually confirmed the absence of a need to account a non-zero injection pulse? Although the HVL with the <ge>IRF did not account all of the variance, it did account 99.99% of it, and there was still enough unaccounted variance for an improvement to occur should this additional parameter be beneficial. The additional injection width term did fit to significance, albeit minimally, although the improvement in the goodness of fit was negligible. That makes sense when the estimate of the injection width of just 0.9 seconds is taken into account. Given this simple experiment, we cannot conclude the delta input assumption to be perfectly true. It is, however, from a practical perspective, very close to true. We can also conclude that whatever it is the once-generalized GenHVL<ge> is fitting in its third moment adjustment, in order to realize the near perfect 2 ppm error, is not associated with inlet effects. We believe, at least with respect to the NLC-type kinetic model, the third moment a4 term in the GenHVL is accounting multiple adsorption sites.
User-Function Fitting of IRFs
As we noted, when you create any user function, PeakLab automatically creates additional UDFs with <ge>, <e2>, <e>, <g>, and <v> IRFs. These are added to the program's internal Fourier fitting of IRF-bearing models. As long as one of these primary IRFs sufficiently models your instrumental distortion, you should not need to fit a specialized IRF user function.
The PeakLab user functions allow you to add any user-defined IRF to the peak model, and to have this processed by the program's internal Fourier processing. You should never need to fit a convolution integral directly in a user function unless you have theoretical data you wish to fit to full machine precision.
In this second part of this tutorial, we will generate an experimental instrument response function and fit a real-world peak to it, and compare it with the <pe> (sum of 1.5 and 1.0 order kinetic decays) IRF.
Before fitting an IRF in a user-defined function, you should be certain the IRF is not already built-in. For the GenHVL and GenNLC family of models, there are a number of experimental built-in IRFs available. These include the half-Gaussian <g>, the sum of order 1 and order 1.5 kinetic decays <pe>, a variable power error model <n> that specializes to the half-Gaussian <g> when the power is 2, and an <s> Student's t that specializes to the half-Gaussian <g> as its adjustable parameter approaches infinity. There is also a variable order kinetic <k> that specializes to the exponential <e> when the order=1. There is also an <l> one-sided generalized normal with a variable log-normal decay.
Here we can also recommend the Search for Optimum Model feature with a data set containing a single peak. The Search for Optimal Model can fit hundreds of potential models in a single unattended step, to data containing a single peak, and it can offer a fit of every ZDD-IRF combination model in the program. You may discover that a specific type of chromatographic peak fits very well with one specific ZDD and IRF pair.
Exploring User-Defined IRFs
While we seek to furnish a broad range of built-in IRFs, in the R&D characterization of peak broadening, you may wish to explore experimental IRFs which are not explicitly built into the program.
A user function can contain a user-defined IRF which is automatically incorporated in the Fourier processing that occurs in the fitting. You can thus fit any IRF as efficiently as the built-in functions. In order to construct an IRF-bearing user-defined peak, you must first define the peak concluding with the Y= expression, as we did above, and then define the IRF concluding with an IRF= expression. You can also create IRFs with more than three parameters. We will note, however, that we have yet to see an instance where an IRF with four or higher parameters was ever fit to significance with real-world data).
For the purpose of this tutorial, we will create a kinetic variant of the <pe> IRF. Second order mass-transfer steps have been postulated as part of the transport mechanisms within a column. The <pe> is a two component IRF that assumes an exponential for the decay with the longest tailing, and a 1.5 order kinetic decay for the narrower component. This 1.5 order decay is used to approximate the complex mix of first and second order mass transfer resistances. There is also a third parameter in the <pe> IRF that specifies the area fractions of the components (by the specification of the area fraction of the narrow width 'p' component).
Click the Select All in the main window. The unselected backgrounds will change from gray to blue. We want to pass all six of the identical replicated data sets to the fitting procedure.
![]() From the Peaks menu, select User Functions... (or use the main toolbar User Function
button). Select UDF 5.
From the Peaks menu, select User Functions... (or use the main toolbar User Function
button). Select UDF 5.
Note the following formula in the main Function field.
Y=GenHVL(x,a0,a1,a2,a3,a4)
IRF1=if(a6==1,exp(-x/a5)/a5,(if(a6<1 && (x>=a5/(1-a6)),0,((2-a6)*pow((a6-1)*x/a5+1,1/(1-a6))/a5))))
IRF2=exp(-x/a7)/a7
IRF=if(x<0,0,a8*IRF1+(1-a8)*IRF2)
The Y= expression creates a five parameter generalized HVL using the GenHVL built-in function. There are then two separate IRFs generated using the formulas for the variable order kinetic and first order exponential IRFs. The IRF= expression sums the two into a final IRF with an a8 area fraction which specifies the area fraction of the narrow-width variable order kinetic component. We have created a nine parameter IRF, but we will lock the power at different values in each of the fits which use this UDF.
We have borrowed the starting estimates for the GenHVL model and set the bounds appropriately. Click OK to exit the UDF procedure.
We will now explore whether a 1.3, 1.4, or 1.6 order kinetic decay is more or less effective than the 1.5 order <pe> IRF built into the program. We will also fit the 1.5 order in the UDF and in the built-in model as a crosscheck.
![]() Click the Local
Maxima Peaks button in the main window (or you can use the Fit Local Maxima Peaks... in
the Peaks menu or main toolbar).
Click the Local
Maxima Peaks button in the main window (or you can use the Fit Local Maxima Peaks... in
the Peaks menu or main toolbar).
We continue to use the remembered settings from the prior fitting in this tutorial.
Click the IRF button. Click the Defaults button. Click OK.
For the primary model, select the GenHVL<ke>-udf5 model.
Right click the apex peak anchor in the first graph, and change the model to GenHVL<pe>. All functions in the program will be shown. The GenHVL<pe> model will be about 35 entries from the top. Click OK (the green check).
Right click the apex peak anchor in the second graph, enter 1.3 in the a6 parameter field and check the lock on a6. Click the OK (the green check).
Right click the apex peak anchor in the third graph, enter 1.4 in the a6 parameter field and check the lock on a6. Cliche the OK (the green check).
Right click the apex peak anchor in the fourth graph, enter 1.5 in the a6 parameter field and check the lock on a6. Cliche the OK (the green check). This user function should give identical results to the built-in GenHVL<pe> model.
Right click the apex peak anchor in the fifth graph, enter 1.6 in the a6 parameter field and check the lock on a6. Cliche the OK (the green check).
We will not lock the order parameter in the last of the replicate data sets. We want to see if the nonlinear optimization can identify a more appropriate order.
Click Peak Fit and check the Fit with Reduced Data Prefit, 2 Pass, Lock Shared Parameters on Pass 1 fit strategy and be sure the Fit using Sequential Constraints (to nudge fits to global minimum when initial estimates weak) is checked. Click OK.
All of six fits should occur within just a few seconds. This example does highlight the value of the built-in Fourier-domain fitting that is done with convolution integrals. Computing the actual integrals, even with the product's quite fast sequential Gaussian quadrature integration algorithm, would take far longer.
Click on the Numeric button of the Numeric Summary is not currently displayed. In the Options menu of the Numeric Summary, choose the Select Only Fitted Parameters
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999836 0.99999834 0.00974889 61,577,025 1.64253940
Peak Type a0 a1 a2 a3 a4 a5 a6 a7
1 GenHVL<pe> 3.98187072 3.04432055 0.04035048 -0.0060416 0.01513660 0.00026115 0.04369642 0.57159048
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999821 0.99999819 0.01016360 56,654,441 1.78525591
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>-udf5 3.97887627 3.04317224 0.04024645 -0.0060492 0.01511583 0.00094236 1.30000000 0.04422617 0.66085047
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999829 0.99999828 0.00993326 59,312,347 1.70525512
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>-udf5 3.97961158 3.04360721 0.04026692 -0.0060470 0.01513193 0.00056408 1.40000000 0.04418319 0.64267159
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999836 0.99999834 0.00974889 61,577,025 1.64253940
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>-udf5 3.98135041 3.04432055 0.04035048 -0.0060416 0.01513660 0.00026115 1.50000000 0.04369642 0.57159049
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999830 0.99999828 0.00992959 59,356,262 1.70399350
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>-udf5 3.98259177 3.04493169 0.04044959 -0.0060359 0.01512540 9.0177e-05 1.60000000 0.04331654 0.40400251
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999836 0.99999834 0.00975441 53,818,965 1.64207643
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>-udf5 3.98121958 3.04426161 0.04034204 -0.0060421 0.01513700 0.00027990 1.49261347 0.04373735 0.58029597
There are a number of items worthy of note. First, all of the powers of this narrow width IRF component give excellent goodness of fits. In order to do this, a8 must adjust the amount of this component rather dramatically. The 1.3 order has a 2/3 area in the overall IRF, the 1.6 order much closer to 1/3.
The optimum does occur very near order 1.5 (the last of the six fits estimates order 1.49 as the optimum).
We have also repeatedly observed that the actual model for the narrow width IRF component in chromatographic peaks is of minor importance. Whether a half-Gaussian in the <ge>, a second narrow width first order exponential in the <e2>, or this 1.5 order kinetic in the <pe>, the model is secondary. An impulse-type function of some form, however, is essential, and the amount of this narrow width component is typically better the half of the overall area of the IRF. We will note our preference for the <ge> IRF when the choice of this narrow width component basically describing an impulse is somewhat arbitrary. Because the tails of the Gaussian are highly compact, there is much less correlation with the higher width component. This means the <ge> IRF is much easier to fit than the <pe> IRF with its much wider tails.
Note the constant value of the higher width exponential component in the IRF. The near-constant value of .043-.044 for the exponential time constant is also seen if the <ge> or <e2> IRF is fitted. We regard this higher width exponential in the IRF as the instrument component since it is close to invariant to system variables. The narrow width component is seen as more of a system component since it will fit to different values as a consequence of sample concentration, temperature, additives, etc.
Click OK in the main Review window to close the Review. You will be asked to save the fits in the data file. Enter IRF <pe> Study for the name and click OK and again click OK to confirm that the fits were saved. You will be returned to the placement screen. Click OK to return to the main window.
Integral IRFs
Before closing this tutorial we will briefly discuss computing convolution integrals directly. To put it succinctly, these are excruciatingly slow. It is likely why true chromatographic models, capable of describing both peaks and instrument effects, have not been published and have not been offered prior to PeakLab in commercial software. You may direct convolution integral fitting useful if you wish to independently validate a built-in PeakLab model, or your own user-defined model where the Fourier IRF processing is used.
The following is a direct convolution integral that is equivalent to user function 5 above:
GFn = GenHVL(x-$,1,a1,a2,a3,a4)
Rsp = a8*RspFn(_RSPFN_K,$,0,a5,a6,0)+(1-a8)*RspFn(_RSPFN_E,$,0,a7,0,0)
FnI = GFn*Rsp
Y = a0*IntQuad(FnI,0,Inf,1e-8)
This is the direct convolution integral that is equivalent to UDF 5, where we used the program's built-in Fourier functionality for IRFs. In this UDF, the peak and IRF are set in a convolution integral. In PeakLab $ is the variable of integration. For chromatographic functions, we recommend x-$ for the peak function, $ for the IRF, and the convolution integral to go from 0 to infinity (Inf). In a direct convolution integral, there is a final Y= user function expression and you must not use the IRF variable for any of the subexpressions in the integral (this will invoke the Fourier IRF processing to add that IRF to the integral). The integrand, here assigned to the variable FnI, must be defined separately and used as the first argument in the integration function.
The IntQuad function is our own very fast successive Gaussian quadrature integration algorithm. Fast is relative, however. The GenHVL<pe>, when fitted to the data set in the last portion of this tutorial, using the same settings, requires about 0.56 seconds on a basic i7 machine. The UDF 5 example above fits in about 1.12 sec with a6 locked at 1.5. The above convolution integral, likewise with a6 locked at 1.5, took nearly 10 minutes for the same count of iterations.
The Fourier IRF processing is capable of an F-statistic of 1E+11 to 1E+14. This is 5.5 - 7 digits of accuracy, appreciably more than any instrument can currently generate. While a full analytic fit can realize an F-statistic up to 1E+32, the 16 digits of machine 64-bit precision, such is seldom of value with real-world data. Note that the above integral, with its 1E-8 eps convergence, can potentially realize at least F=1E+16, 8 or more digits, but at a steep computational price, in this case a fit that is over 500 times slower.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999836 0.99999835 0.00972673 61,857,982 1.63507907
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>Int-udf6 3.98174681 3.04409804 0.04035241 -0.0060417 0.01514828 0.00014762 1.50000000 0.04370534 0.69524754
The above are the results from fitting this direct convolution integral. While there are differences in the parameters, the goodness of fit is only ever so slightly better, 1.635 ppm error vs. 1.642 ppm. The reduction in the a5 1.5 order time constant, and its increase in area, fits with the latitude one has in accommodating the narrow width component of the IRF. Again we note that the <pe> with a 1.5 order kinetic is harder to fit because of the greater correlation between the IRF components. The <ge> is much easier to fit because the components are much more independent. We will note that the genetic procedure in the IRF Deconvolution preprocessing does not have this limitation. There you may well see that the <pe> is much closer to the theoretically correct IRF, especially with tiny porous media with high mass transfer resistances.
Practical Considerations
Fitting an integral convolution, even using the fast IntQuad function, can invoke up to 9000 evaluations of the user function integrand per data point, per iteration. To make such fitting tolerable, we suggest limiting integral-bearing fits to experimental studies, that you use only a single peak, and that you limit that peak's data to no more than about 500-1000 points.
Especially within integrations, you should use built-in functions wherever possible when constructing a user function. A great many of PeakLab's models, and many IRFs, are not defined at all x-values and as fully compiled functions will run faster than your putting in the formulas and seeking to manage the undefined regions. You should only enter a peak model's formula or a response function model's formula, if these are not built into the product.
Avoid Singularities, Discontinuities, and Non-Smooth Integrands
In fitting chromatographic convolution models, the IRFs begin at a maximum at t=0 and decay as t increases. If you are fitting the convolution integrals directly, you should design the convolution integral to avoid the discontinuity that would exist in the standard definition of a convolution integral whose limits go from -infinity to +infinity. Even if you use an IF or Heaviside function, it will be better to change the limits of integration than to attempt to integrate across such a discontinuity. The algorithms often manage it, but at the price of appreciably slower integrations.
Integral Algorithms
The Gaussian quadrature, the IntQuad() function, is usually the fastest of the integration procedures. The adaptive quadrature, the IntAdapt() function, will use up to 10,000 evaluations of the user function integrand per data point, per iteration, and is generally slower. The Romberg algorithm, the IntRomb() function, can use many more, and it is generally the slowest of the three procedures. We recommend the IntQuad() as the principal algorithm, the IntAdapt() as the first fallback, and the IntRomb() as the final fallback. You must select which integration function is to be used when you create an integral-bearing user-defined model.
If we fit this same data to UDF 6, except with IntAdapt substituted for the integration function, the fit requires 35 minutes as compared to the 10 minutes or so for the Gaussian quadrature procedure.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999836 0.99999835 0.00972672 61,858,042 1.63507749
Peak Type a0 a1 a2 a3 a4 a5 a6 a7 a8
1 GenHVL<ke>Int-udf7 3.98174680 3.04409805 0.04035241 -0.0060417 0.01514828 0.00014762 1.50000000 0.04370534 0.69524754
At this point, the differences in the goodness of fit and parameter values are negligible when contrasted with the IntQuad fit.
If we fit this same data to UDF 6, except with IntRomb substituted for the integration function, the fit requires close to 2 hours!
Note that all three of the integration functions have a fourth argument where the eps is specified. We typically use 1e-8 for well-behaved integrands. For especially difficult models, you should be able to specify 1e-5 or 1e-4 and still realize good fits with real world data, and fitting times should be improved.
Convergence Errors
During fitting, PeakLab disables the convergence error handling. It is rare that these integration algorithms will fail to converge, and when it does occur, the errors are often close to the target epsilon. Note that the eps in the integration routines is a target only. For example, if you fit a 1000 point single peak data set to a high parameter count UDF where 100 iterations are required, there can be a million or more integrations. You should not assume each of these converged to the specified eps. A fit is not terminated or interrupted when a failed convergence occurs. Such most often occurs at extreme bounds or when parameters temporarily assume troublesome values for a given iteration, neither of which impact the final fit. When you see a suffix such as <2> in the Iter field displayed during fitting, this means two of the user-function parameter constraints were imposed within the current iteration.
Exploring Complicated Peaks and IRFs
All integral UDFs will include variants with the built in <ge>, <e2>, <g>, <e>, and ,v> IRFs, and these will use the fast Fourier IRF processing adding very little additional computation time to that of the user function. You can thus construct a peak which is itself a convolution integral and fit that model with one of these standard kinetic, probabilistic, or spectral IRFs with no additional effort. If you need a custom IRF with such a convolution integral user-defined peak, the construction of the UDF is straightforward:
Fn=Gauss(x-$,1,a1,a2)
Rsp=RspFn(_RSPFN_E,$,1,a3,0,0)
FnI=Fn*Rsp
Y=a0*IntQuad(FnI,-Inf,Inf,1e-8)
IRF=RspFn(_RSPFN_K2,x,0,a4,a5,0)
In this user function example, a Gaussian is convolved with a two-sided exponential response function to produce a core peak smeared to each side by an equal magnitude first order kinetic distortion. That peak is then further distorted by a variable order one-sided kinetic IRF. Note the 1 in the RspFn for a centered two-sided response function and the -Inf to Inf limits on the integration. Note also the use of the $ variable of integration in the peak and its IRF in the core convolution integral, and the use of just the variable x in the IRF where this Fourier convolution is implicit. Note also that the x for the Fn is the actual x of the data, and the x for the IRF is the delta x from 0 for the response function.


 |