PeakLab v1 Documentation Contents AIST Software Home AIST Software Support
HPLC Gradient Peaks - Fitting Unwound Data (Tutorial)
Unwinding the Gradient and Fitting a Once-Generalized Model with or without an IRF
Once you have determined the IRF model and parameters for a gradient, you can use this system identification of the gradient to "unwind' the gradient as if an isocratic elution had occurred for all peaks. In this case, there is a preprocessing step of unwinding the gradient and then a subsequent fitting of the peaks with any one of the conventional isocratic models, including those with IRFs to manage any portion of the IRF which survived the gradient.
The process of using convolution to 'unwind' the gradient for subsequent conventional isocratic fitting is also discussed in the HPLC Gradient Peaks topic.
![]() Use the File menu's Open item and select the file HPLCGradientStd02.pfd from the
program's installed default data directory (\PeakLab\Data).
Use the File menu's Open item and select the file HPLCGradientStd02.pfd from the
program's installed default data directory (\PeakLab\Data).
The file contains eight data sets, each a baseline-corrected data set from a single standard which elutes during the gradient. This data was used in the first gradient peak tutorial, where peaks were fitted to their compressed shapes, and in the second gradient peak tutorial, where the gradient was fitted and deconvolved to internally produce an estimate of a constant mobile phase peak. In this third gradient peak tutorial we will actually generate and fit these gradient deconvolved shapes.
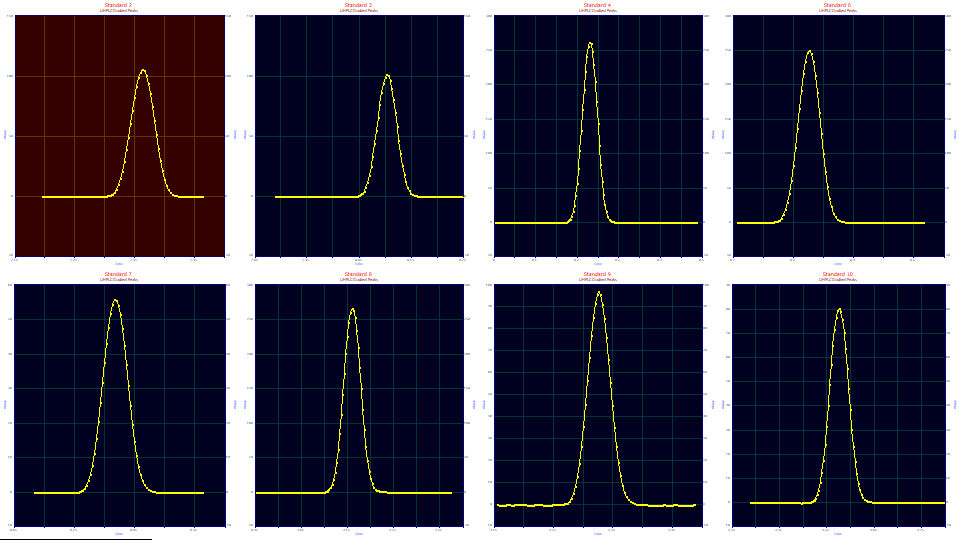
Unwinding the Gradient with a Convolution Model
In the second gradient peak tutorial, we estimated a half-Gaussian IRF to unwind the gradient as having an SD of .0203. We will use this value to create a data level consisting of unwound data. We also found that one-step deconvolution models, with no IRF, produced an average unaccounted variance of 5.4 ppm. We want to see stronger fits in this two-step process.
We will now use the IRF Deconvolution procedure in reverse. We will employ an IRF as a convolution model to undo the compression that occurs within the gradient. We will use the estimation of the system gradient model from the prior fits to unwind the gradient's compression of the peak shapes.
Click on the IRF Deconvolution button and ensure the following settings are selected:
Response Fn
<g> Gaussian
Convolve Right (we want convolution, not deconvolution)
IRF Parameters
0.0203 (the
value from the GenHVL<*g*> deconvolution integral fit
Genetic Alg Optimization
These settings will not be used
Fourier Filter
None (no filtering is generally needed for convolution)
Post-Processing
Zero Baseline Points - unchecked (baseline will be
actual subtracted values)
Zero Negative Points - unchecked (negative
baseline points are permitted)
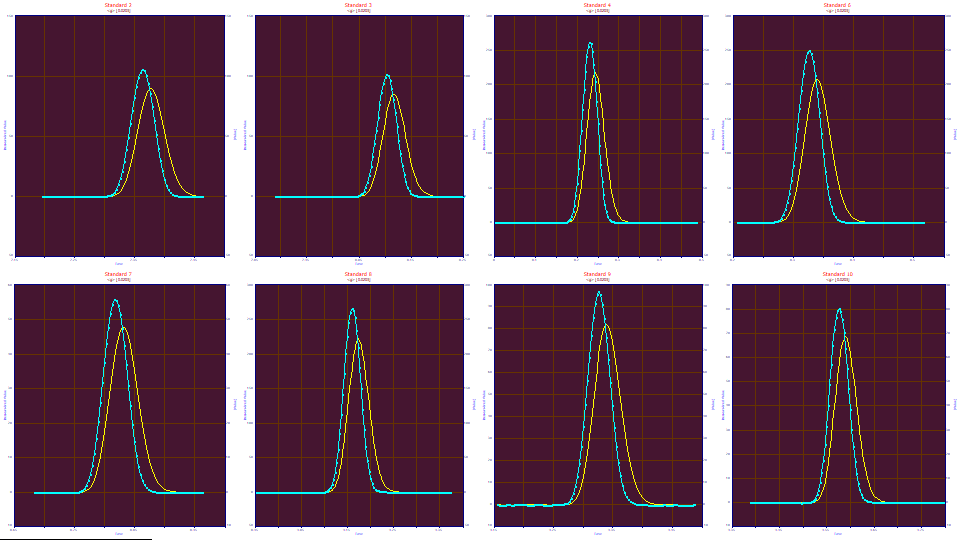
Note the loss of resolution between the blue gradient peaks, and the yellow peaks after the gradient has been unwound.
Click OK to close the procedure and OK to accept the default name and data set titles for the new data level.
Fitting the Unwound Gradient Peaks to the GenHVL<ge> Model
If we successfully unwound the gradient compression in the convolution, we should be able to treat the peaks as having occurred in an isocratic elution.
Click the Local Maxima Peaks button in the main window to open the peak placement screen. We will use the same settings as in the first gradient fitting tutorial where an isocratic peak was fitted:
Peak Detection
Set Sm
n(1) to 5
Peak Type
Select Chromatography
in the first dropdown
Select GenHVL <ge> as the model in the
second dropdown
Scan
Set the Amp %
threshold to 1.5 %
Leave Use Baseline Segments unchecked
Be sure Use IRF,ZDD is checked
Vary
Leave width a2
and shape a3 checked, all other unchecked
Click the IRF button, and if needed set the defaults for the <ge> IRF as 0.004 for the g width (sd), 0.016 for the e width (tau), and 0.625 for the g area fraction. Check the Lock for the g area fraction. This is what we previously did with the isocratic peak to ensure significance of the fitted parameters. We want to use proportionately lower starting estimates than the defaults to account the portion of the IRF that disappears in the gradient and is not unwound. Click OK.
Click Peak Fit to open the fit strategy dialog. Select the Fit with Reduced Data Prefit, 2 Pass, Lock Shared Parameters on Pass 1. Leave the Fit using Sequential Constraints box checked. Click OK. Once again, if you are fitting any peak model bearing an <irf>, you will almost always use this 2-pass option where the main peak parameters are first estimated while the starting estimates for the IRF and ZDD parameters are held constant.
Click Review Fit. You will see some exceptional fits. Four of the eight have fit errors less than 1 ppm unaccounted variances, an r2 coefficient of determination of .999999 or higher.
Be sure As Fitted is selected in the first dropdown. This will be the unwound data and for most color schemes, a white curve.
Select the IRF Deconv. option in the second dropdown. This plots, in the first aux component color (red by default for most color schemes), the fitted peak with the IRF removed, the GenHVL, the peak you would see if there existed no system/instrumental distortion. The difference between the red and the white curves is the instrumental distortion recovered in the unwinding of the gradient.
Change the third dropdown to Partial Deconv. The second auxiliary component color is usually green by default. For this model, the green peak (the partial deconvolution) is the pure HVL, the theoretical peak after the ZDD's third moment non-idealities are mathematically removed. The red curve remains the peak absent the system/instrumental effects, the GenHVL. The difference between the red and green curves is that which occurs from fitting an asymmetry or additional third moment parameter in the ZDD, the zero distortion density.
Change the fourth dropdown to Full Deconv. The third auxiliary component color is usually blue by default. For this GenHVL<ge> model, the blue peak (the full deconvolution) is the pure Gaussian, the peak that would exist at infinite dilution. Despite the low concentrations, the a3 intrinsic chromatographic distortion changes the peak shape from the infinite dilution blue peak, to the fronted HVL peak in green.
![]() Click the Hide Y2 plot in the graph's toolbar.
Click the Hide Y2 plot in the graph's toolbar.
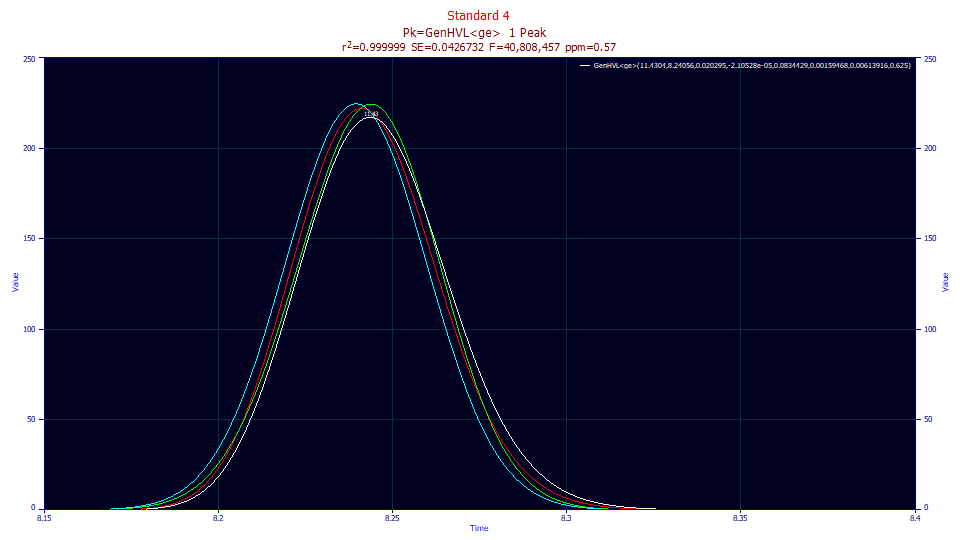
Double click the third graph (the one with the best goodness of fit) and use the mouse to zoom in the peak.
For reference, this is the isocratic peak fitted in the first of the gradient tutorials.
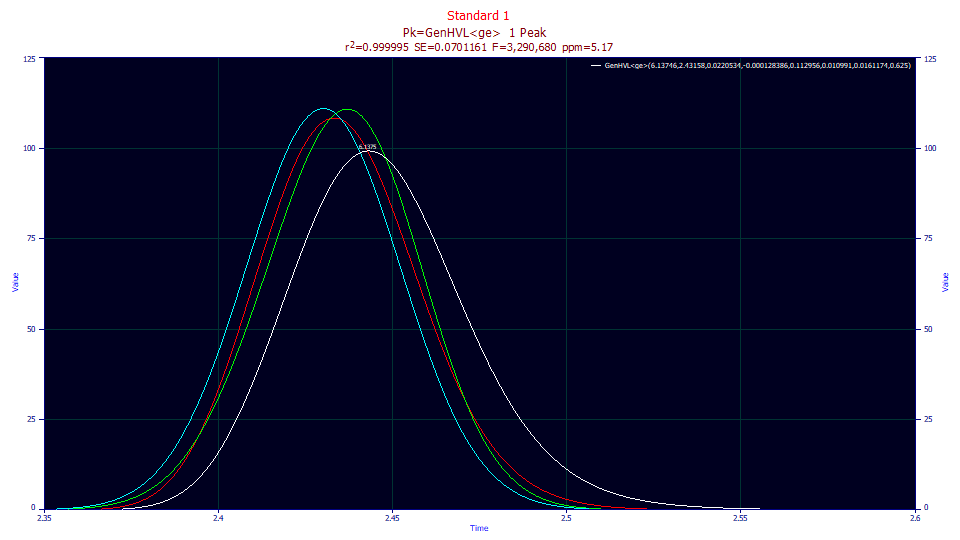
Although the different deconvolutions are more pronounced in fit of the the isocratic peak (a significant part of the IRF is destroyed in the gradient), those of the unwound gradient follow the same pattern. The a3 fitted chromatographic distortion remains fronted (a3<0).
Double click the graph to restore all plots. Click on the Numeric button to open the numeric summary and under the Options menu, choose Select Only Fitted Parameters and then check Measured Values, Deconvolved Moments, and Average Multiple Fits. Scroll to the bottom of the report to see the averages.
We'll first compare the separate isocratic fit in tutorial 1 alongside the averages from these unwound gradient fits to see how much of the asymmetry in the ZDD, and how much of the IRF widths, are present in the unwound or decompressed peaks.
Peak Type a0 a1 a2 a3 a4 a5 a6 a7
Isocratic GenHVL<ge> 6.13746193 2.43158324 0.02205342 -0.0001284 0.11295617 0.01099098 0.01611737 0.62500000
Unwound GenHVL<ge> 6.90040719 8.62110363 0.02097263 -2.879e-5 0.09334915 0.00520456 0.00497757 0.62500000
The a6 exponential width recovered from the unwound gradient peaks is only about 30% of the isocratic peak's value, very close to our expectation in terms of that portion of the IRF surviving the gradient we estimated by the genetic algorithm optimizations that were made in the first tutorial. For the a5 half-Gaussian component of the 2-component IRF, we see about 50%, and for the a4 ZDD asymmetry, we see about 83% of the isocratic value. Because the values are smaller, closer to zero, there are significance issues with the 2-component IRF in many of the fits.
Average for 8 Fits
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999770 0.99999751 0.04037905 18,424,491 2.30209573
Peak Type a0 a1 a2 a3 a4 a5 a6 a7
1 GenHVL<ge> 6.90040719 8.62110363 0.02097263 -2.879e-5 0.09334915 0.00520456 0.00497757 0.62500000
Measured Values
Peak Type Amplitude Center FWHM Asym50 FW Base Asym10
1 GenHVL<ge> 127.569310 8.62662338 0.05154018 1.04872096 0.10302937 1.13018965
Peak Type Area % Area Mean StdDev Skewness Kurtosis
1 GenHVL<ge> 6.90027137 100.000000 8.62844834 0.02197493 0.21018122 3.10351136
Deconvolved Moments
Peak Type Area Mean StdDev Skewness Kurtosis Amplitude Center
1 GenHVL 6.90028425 8.62444886 0.00045822 0.20042433 3.06996218 131.172976 8.62282512
Peak Type Area Mean StdDev Skewness Kurtosis Amplitude Center
1 HVL 6.90039888 8.62440603 0.00044199 -0.0781386 3.00154004 133.023648 8.62577527
Peak Type Area Mean StdDev Skewness Kurtosis
1 Gauss 6.90040235 8.62011954 0.00044018 -7.762e-5 2.99970243
We see what we expect, the kurtosis no longer showing the gradient compression observed in the second gradient tutorial.
Click OK to close the Review, OK to accept a saved GenHVL<ge> fit in the data file, and OK to acknowledge the revision in the file. You will be back in the placement screen.
Fitting the Unwound Gradient Peaks to the Gen2HVL<e> Model
Here we will make the assumption that the remaining accuracy that can be realized in fitting must come from accepting that certain peaks were unwound too strongly and others too weakly in using a common convolution for the unwinding of all of the peaks. We will thus fit the Gen2HVL<e> model to allow the power of the decay to also be fitted, and we will also fit only the simple <e> exponential since the half-Gaussian component of the <ge> IRF cannot be consistently fitted to significance. We want the goodness of fit further improve. We also want the power of decay parameter to be a crosscheck on the unwinding, and we would like to see all parameters significant in all of the fits. We will also lock the exponential value to ensure that there is no interaction with the power of decay parameter.
Select Gen2HLC <e> as the model in the second dropdown.
Click IRF and set the <e> e width (tau) to 0.005 and check the Lock box. Click OK.
Click Peak Fit to open the fit strategy dialog. Be sure the Fit with Reduced Data Prefit, 2 Pass, Lock Shared Parameters on Pass 1 and Fit using Sequential Constraints boxes are checked. Click OK. Click Review Fit when the fits are complete.
If needed, click on the Numeric button to open the numeric summary, and once again scroll to the bottom of the report to see the averages.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999892 0.99999885 0.03617719 32,653,143 1.07749161
Peak Type a0 a1 a2 a3 a4 a5 a6
1 Gen2HVL<e> 6.89838721 8.62079134 0.02098648 -2.581e-5 2.00640831 0.09061789 0.00500000
Measured Values
Peak Type Amplitude Center FWHM Asym50 FW Base Asym10
1 Gen2HVL<e> 127.554469 8.62661407 0.05157938 1.05009849 0.10295706 1.12993439
Peak Type Area % Area Mean StdDev Skewness Kurtosis
1 Gen2HVL<e> 6.89820516 100.000000 8.62842884 0.02193464 0.20550638 3.06967023
Deconvolved Moments
Peak Type Area Mean StdDev Skewness Kurtosis Amplitude Center
1 Gen2HVL 6.89831839 8.62378304 0.00045505 0.19903353 3.06480468 131.146189 8.62215840
Peak Type Area Mean StdDev Skewness Kurtosis Amplitude Center
1 HVL 6.89838134 8.62375689 0.00044222 -0.0705598 3.00119686 133.010735 8.62498612
Peak Type Area Mean StdDev Skewness Kurtosis
1 Gauss 6.89838353 8.61983841 0.00044085 -6.149e-5 2.99976007
The eight fits now average just above 1 ppm unaccounted variance. Note that no parameters are grayed; every parameter is significant. Further, the a4 power of decay averages to 2.006, a confirmation of the successful unwinding of the gradient. The a4 varied from 1.981 to 2.037 suggesting the peaks did experience modest differences in the unwinding of the gradient, which in turn implies the possibility of differences in the compression in the original gradient peaks.
Click OK to close the Review, OK to accept a saved Gen2HVL<e> fit in the data file, and OK to acknowledge the revision in the file. You will be back in the placement screen.
In Perspective Once Again
In the third IC tutorial, we discussed the expectations in goodness of fit in prior versions of the peak fitting technology. If we separate the three fronted cation peaks in the data used in these IC tutorials and fit them with the same diligence we gave to the gradient peaks in this tutorial, we will see the following ppm unaccounted variance averages for the sixteen different conditions of concentration and additive level:
|
Peak |
HVL |
GenHVL |
HVL<ge> |
GenHVL<ge> |
|
Li+ |
4779 |
4660 |
225.25 |
3.99 |
|
Na+ |
1657 |
1532 |
36.42 |
2.16 |
|
NH4+ |
966 |
830 |
17.91 |
1.82 |
The HVL column represents the best fits attainable in the prior v4 of this peak-fitting technology. The GenHVL model includes the ZDD asymmetry adjustment but not the IRF adjustment. The HVL<ge> includes the IRF adjustment, but not the ZDD. The GenHVL<ge> includes both the IRF and ZDD adjustments. As we have stressed in the first six tutorials, the IRF and the higher moment(s) ZDD are exceptionally important in realizing close to zero error fits.
There was a point where sub-100 ppm unaccounted variance fits (r2=0.9999) were deemed impossible using all known theoretical and empirical models. Fitting the IRF and ZDD components in a generalized HVL or NLC model have made that level of fitting routine in PeakLab. As such, our objectives with fitting have changed. We now want to estimate peaks to such a high degree of accuracy that any diminished separation performance will not only be quantified, but where it can be easily observed to change across time.
In seeing these isocratic fits of these high S/N peaks fitted to 2-4 ppm, and with our last set of unwound gradient peaks fitting to 1 ppm, we can note that gradient peaks are capable of seeing equivalent goodness of fits.
Comparing the Three Approaches
|
Model |
a0 |
a1 |
a2 |
a3 |
a4 |
a5 |
a6 |
a7 |
F-stat |
ppm uVar |
|
(1) Gen2HVL |
6.89712 |
8.61235 |
0.01836 |
-6.535E-06 |
2.13182 |
0.00782 |
|
|
1,259,491 |
16.66 |
|
(2) GenHVL<*g*> |
6.89667 |
8.62628 |
0.02142 |
-2.809E-05 |
0.09236 |
0.02032 |
|
|
11,487,966 |
5.38 |
|
(2) GenHVL<*k*> |
6.89733 |
8.62310 |
0.02081 |
-3.058E-05 |
0.09493 |
0.02122 |
0.71312 |
|
5,759,968 |
5.70 |
|
(3) GenHVL<ge> |
6.90041 |
8.62110 |
0.02097 |
-2.879E-05 |
0.09335 |
0.00520 |
0.00498 |
0.62500 |
18,424,491 |
2.30 |
|
(3) Gen2HVL<e> |
6.89839 |
8.62079 |
0.02099 |
-2.581E-05 |
2.00641 |
0.09062 |
0.00500 |
|
32,653,143 |
1.08 |
While there are the differences we covered in the second gradient tutorial with respect to (1) fitting the eluted compressed gradient shape and (2) fitting a deconvolved model that internally unwinds the gradient, here we see no difference in a0-a3, the principal HVL parameters, nor in a4, the skewness adjustment to the zero distortion density, between (2) fitting a deconvolved model that internally unwinds the gradient and (3) fitting the convolved (unwound) data.
Direct Closed Form Fit - Fitting the Compressed Shapes
In the direct fits of the Gen2HVL and GenHVL[Q] to the gradient shapes (the first gradient tutorial), we see compressed a2 widths, much smaller a3 distortions, a much higher a4 power of decay, and an a5 ZDD asymmetry that is also a great deal smaller. The gradient shape is fitted, and the a4 power of decay parameter directly maps the strength of the gradient. The fits are slightly weaker, though still very effective. An isocratic model is being fitted to a gradient shape, but the variable power of decay, which fits the reduced kurtosis from the compression, the thinning of the tails, allows for these models to be effective with gradient shapes. With respect to the HVL estimated parameters, the a1 location will not match exactly, the a2 deconvolved Gaussian width will be smaller, and the a3 chromatographic distortion will be far smaller, when contrasted with the isocratic deconvolved values for the peaks. The advantages of this approach are the direct measurement of the gradient strength and the ease of fitting very fast closed form models.
If you have no need to know what the peak would actually look like in an isocratic elution, and you need little more than the areas of some count of overlapping or hidden peaks, or if you want the plate count to include the gradient's contribution to the separation, you should fit the compressed shapes directly. You give up a small measure of accuracy (very small when seen in context), the fitted HVL peak widths will reflect the gradient compression, and the theoretical plates, at least by the method of moments, should reflect the separation that actually took place. You will still get a measure of the gradient strength in the fourth moment term. It is indirect, a measure of the fourth-moment tailing, but it was, at least for us, an effective estimate of the strength of the gradient.
Fitting the Gradient with a Deconvolution Model
In the GenHVL<*g*> and Gen2HVL<*g*> deconvolution models, we fit an intrinsic unwinding of the gradient. The a0-a3 HVL parameter values represent a best-case scenario where each peak is estimated as unwound, even if the data itself does not see this unwinding in an extrinsic step. The GenHVL<*g*> forces estimates which produce a full unwinding, and the half-Gaussian width measures the SD used for that unwinding. The Gen2HVL<*g*> additionally fits a power of decay which confirms the unwinding. The advantages of this approach are isocratic peak parameter estimates, and better goodness of fits since the gradient is modeled. The disadvantages include the need for baseline-resolved peaks with a strong baseline on each side of each peak, the need for a strong zero baseline on both sides of the data for the Fourier continuity, the need for a well-designed Fourier noise filter, and the fitting which must occur in the Fourier domain will be much slower, and prone to local minima very near the global minimum as a consequence of this noise filtration.
In this approach the estimate of the power of the gradient is in the unwind parameter(s), not in a parameter that describes the thinness of the tails of the gradient shape.
Because of these drawbacks, an approach where the gradient is fitted only on single peak standards used to test system stability is recommended. One can fit a deconvolution model to production data with many peaks, and we will do so at the end of this last gradient tutorial, but it is not, at least at present, a viable analytic approach for production data, since the requirements mentioned above will often be lacking.
Convolution Unwind and Subsequent Isocratic Fit
Although we need to know the IRF parameter(s) for the gradient, the unwinding occurs in a forgiving convolution. In this tutorial, we fit the GenHVL<ge> and Gen2HVL<e> models, the latter offering an incredible 1 ppm average unaccounted variance fitting accuracy. The GenHVL<*g*> of the one-step deconvolution fitting, and the Gen2HVL<e> of the unwound convolved data produce essentially the same HVL parameter estimates for an unwound isocratic peak. The advantage of the two step procedure is clearly the exceptional goodness of fits. The disadvantages are the need to predetermine the IRF for the gradient and the loss of resolution arising from the convolution of the data in the unwinding, a broadening that may make certain fits more difficult if a local maxima peak is lost or becomes less well-defined. There is also the issue of each peak seeing exactly the same unwinding in this preprocessing step, where the first two approaches process the gradient on a peak by peak basis.
If you compare the goodness of fit values from the gradient peak fits with the above cation fit reference values, they are comparable only when you make the extra effort to fit the gradient or to unwind the estimated gradient prior to fitting. Such may well be worthwhile, especially with critical certification of systems. To have everything mapped to within 1ppm unaccounted variance does mean you will catch any anomaly swiftly and you are effectively measuring everything that can be modeled. You are also getting an estimate of the true peak and its key parameters with the impact of the gradient removed. You see very accurate deconvolved Gaussian widths and chromatographic shapes from the a2 and a3 values, and when fitting a fourth moment Gen2 model, you also get a confirmation of the unwinding of the gradient, and such is available on a peak-by-peak basis if you have strong baseline resolved peaks and if this a4 power of decay parameter is not shared across the peaks.
Importing Gradient Peak Production Data
![]() Use the File menu's Open item and select the file HPLCGradientPeaks.pfd from the
program's installed default data directory (\PeakLab\Data). The file contains one gradient peak data set
which has already been preprocessed for baseline.
Use the File menu's Open item and select the file HPLCGradientPeaks.pfd from the
program's installed default data directory (\PeakLab\Data). The file contains one gradient peak data set
which has already been preprocessed for baseline.
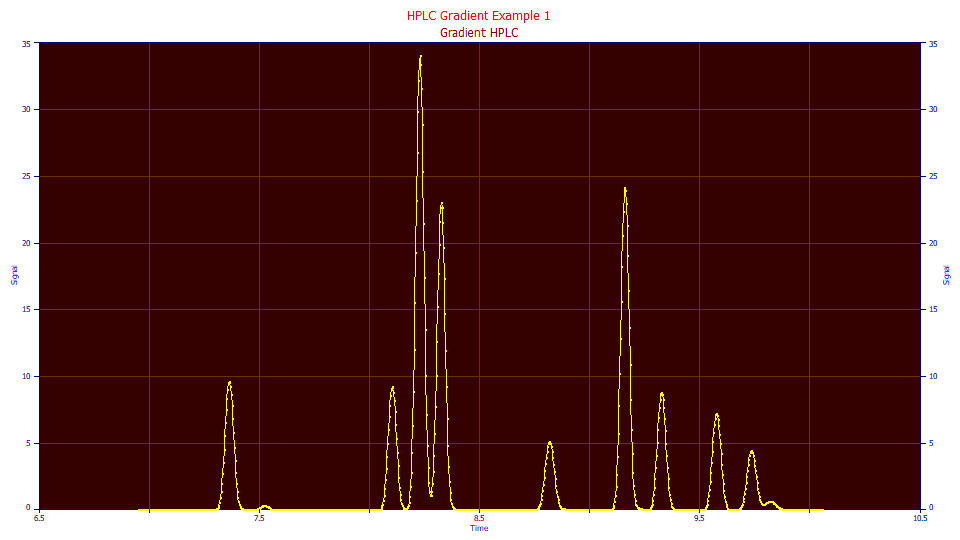
Click on the IRF Deconvolution button and ensure the following settings are selected:
Response Fn
<g> Gaussian
Convolve Right
IRF Parameters
0.0203
Genetic Alg Optimization
These settings will not be used
Fourier Filter
None (a frequency domain filter is not normally required
for convolution)
Post-Processing
Zero Baseline Points - unchecked (baseline will be
actual deconvolved values)
Zero Negative Points - unchecked (negative
baseline points are permitted)
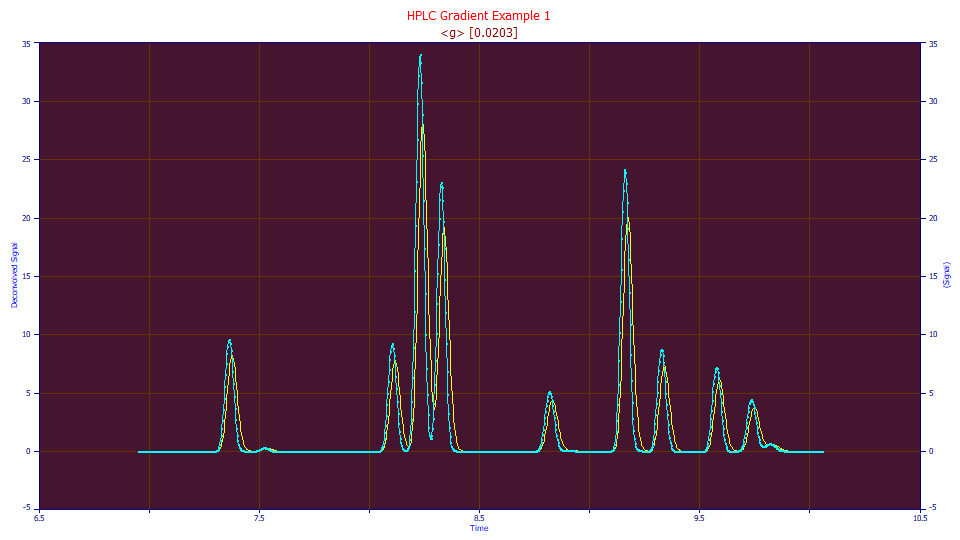
The blue curve is the compressed data as eluted, the yellow curve is the convolution of the data and estimated gradient, the 'unwound' data. Note that the last peak loses its local maximum (there is no longer a defined apex) as it merges into the larger adjacent peak.
Click OK. Click OK to accept the default titles. There is now a second data level in the file with the name DC(<g>,CR,0.0203,None,0). The default data levels names reflect the global settings in the procedure. This level originated with the IRF Deconvolution procedure (DC), with the half-Gaussian IRF (<g>), a right-side convolution (CR), the <g> parameter (0.0203), and with no filter (None,0). The actual settings, including all custom settings, will appear in the automatically generated second graph title. In this case, the graph title is identical:
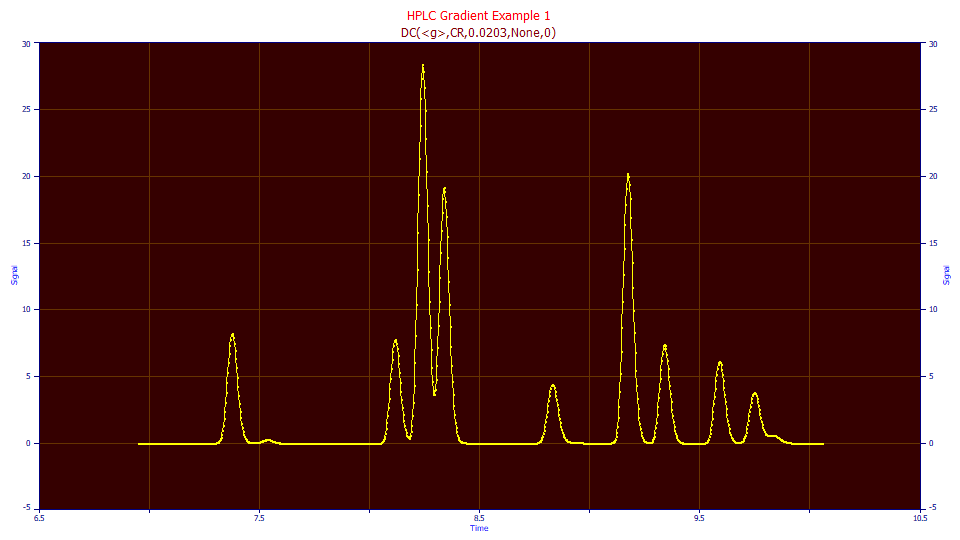
Fitting the Gradient Peak Shapes Directly
Select Imported Data from the Data Levels dropdown.
Click the Local Maxima Peaks button in the main window to open the peak placement screen. We will use the same settings as in the first gradient fitting tutorial for the isocratic peak:
Peak Detection
Set Sm
n(1) to 5
Peak Type
Select Chromatography
in the first dropdown
Select Gen2HLC as the model in the second
dropdown
Scan
Set the Amp %
threshold to 0.5 % (we need to catch the tiny peak)
Leave Use Baseline Segments unchecked
Be sure Use IRF,ZDD is checked
Vary
Leave width a2
and shape a3 checked
Check a4 (we want the power of decay to vary
on a peak by peak basis to estimate the gradient present at each peak)
We do not have to worry about the IRF since we are not fitting one. You should see 11 peaks automatically placed, all local maxima peaks (an apex is seen in the data).
Click Peak Fit to open the fit strategy dialog. Select the Fit with Reduced Data Prefit, Cycle Peaks, 2 Pass, Lock Shared Parameters on Pass 1. Leave the Fit using Sequential Constraints box checked. Click OK. The Cycle Peaks strategy performs certain fits on a peak by peak basis to ensure the fitting accuracy of small peaks. You will almost always want to use this strategy when multiple peaks are present. Despite the complexity of the Gen2HVL closed form model, the fitting should require less than a second, all cores of your CPU utilized for the fitting.
Click Review Fit. We have a lovely 19.97 ppm unaccounted variance.
![]() If needed, click Show or Hide Legend Titles in the graph toolbar to shift the peak formulas to
the left of the graph's area.
If needed, click Show or Hide Legend Titles in the graph toolbar to shift the peak formulas to
the left of the graph's area.
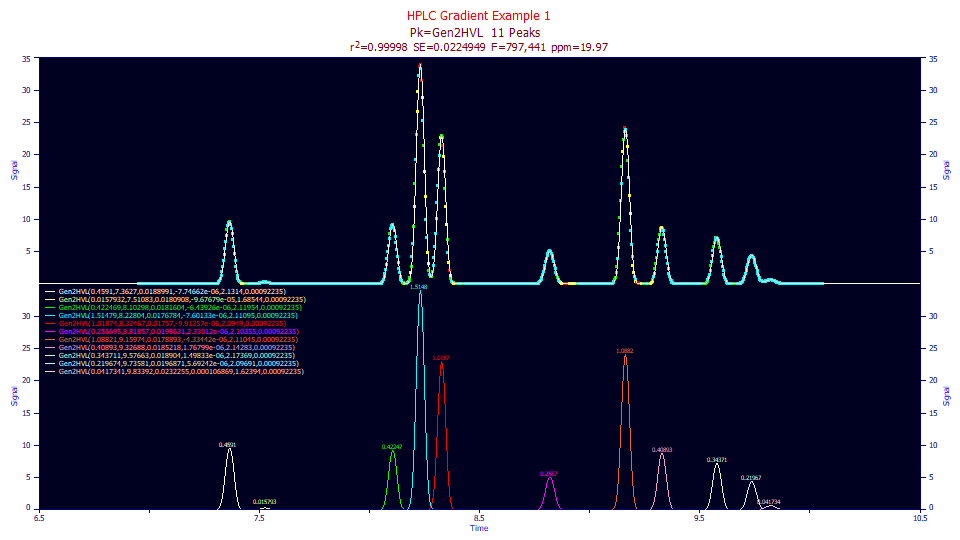
Click on the Numeric button to open the numeric summary and under the Options menu, choose Select Only Fitted Parameters.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99998003 0.99997875 0.02249492 797,441 19.9725764
Peak Type a0 a1 a2 a3 a4 a5
1 Gen2HVL 0.45909985 7.36270241 0.01889909 -7.747e-06 2.13140190 0.00092235
2 Gen2HVL 0.01579321 7.51083120 0.01809079 -9.677e-05 1.68544369 0.00092235
3 Gen2HVL 0.42246880 8.10297608 0.01816043 -6.439e-06 2.11953543 0.00092235
4 Gen2HVL 1.51479133 8.22803626 0.01767838 -7.601e-06 2.11094630 0.00092235
5 Gen2HVL 1.01874016 8.32467112 0.01756996 -9.913e-06 2.09490368 0.00092235
6 Gen2HVL 0.25669538 8.81856822 0.01986309 2.3301e-06 2.10354569 0.00092235
7 Gen2HVL 1.08821191 9.15973955 0.01788931 -4.334e-06 2.11045430 0.00092235
8 Gen2HVL 0.40892996 9.32687815 0.01852180 1.768e-06 2.14282935 0.00092235
9 Gen2HVL 0.34371131 9.57662518 0.01890399 1.4983e-06 2.17368641 0.00092235
10 Gen2HVL 0.21967402 9.73581480 0.01968705 5.6924e-06 2.09690879 0.00092235
11 Gen2HVL 0.04173413 9.83391853 0.02322546 0.00010687 1.62394283 0.00092235
The a5 fails significance testing but again this tests for differences from zero. In this case, a tiny positive a5 simply means the fit was unable to find a statistically significant third moment variation from the the HVL's symmetric Gaussian zero-distortion density assumption (a5=0).
The a4 confirms the thinning of the tails on all of the peaks of significant area. The two very small peaks either lacked sufficient information, or the baseline correction introduced a sufficient error, so that the gradient compression could not be fitted. Also, when peaks are overlapped, as occurs here, this fourth moment may not have the benefit of baseline on either side. As you will note, however, except for the smaller peaks, all fit close to the 2.13 of the averaged standards. We know, in fitting the actual gradient shape, we will have a lower a2 and lower magnitude a3 chromatographic distortions. Here two of these are indistinguishable from zero (neither tailed nor fronted) and three fit to tiny positive (tailed values).
Click OK to close the Review, OK to accept a saved Gen2HVL fit in the data file, OK to acknowledge the revision in the file, OK to return to the placement screen, and OK one last time to return to the main window.
Fitting the Unwound Gradient Peaks
We know that most of the isocratic IRF is effectively offset in the gradient, and that all that remains (that we can identify) in the unwound gradient peaks is a small amount of residual exponential distortion. For this ultra HPLC reverse phase chromatography, we found that an isocratic peak only had about 1/3 of the exponential IRF width in comparison with high S/N IC peaks (about 0.016 as contrasted with 0.040-0.045). Further, we found only about 0.005-0.006 of this exponential IRF survives in a gradient peak.
In our experience, fitting this small of an IRF is probably not going to add a significant benefit, and it will generally be simpler, and far faster, to fit a closed form model, typically the GenHVL, or the Gen2HVL (if you wish to fit a fourth moment term to confirm the successful unwinding of the gradient). You will wish to fit peaks with and without this <e> IRF to be certain, but in our experience a simple closed form model suffices unless you are seeking to estimate this small residual exponential IRF.
Select the uppermost 'DC' data from the Data Levels dropdown.
Click the Hidden Peaks - Second Derivative button in the main window to open the peak placement screen. We know the last of the peaks is now hidden. We will see if the D2 algorithm can automatically place this peak for us.
Baseline
No Baseline
Peak Detection
Set Sm
n(1) to 5 (this small
level of D2 smoothing is possible because the S/N is very good on this data)
Peak Type
Select Chromatography
in the first dropdown
Select GenHVL as the model in the second dropdown
Scan
Set the Amp %
threshold to 0.5 % (to again catch the tiny peak)
Leave the D2 % threshold at 0.00 (all smoothed
D2 minima drop below 0)
Leave Use Baseline Segments unchecked
Be sure Use IRF,ZDD is checked
Vary
Leave width a2
and shape a3 checked (the widths and shapes must vary for
each peak)
Leave a4 unchecked (the third moment deviation
from non-ideality will be shared by all peaks)
You should see 11 peaks, all automatically placed.
We will not be fitting an IRF, and unless you altered the ZDD default starting estimates for the GenHVL, there is no need to open the ZDD dialog.
Click Peak Fit to open the fit strategy dialog. Be sure the Fit with Reduced Data Prefit, Cycle Peaks, 2 Pass, Lock Shared Parameters on Pass 1 and Fit using Sequential Constraints boxes are checked. Click OK.
Click Review Fit when the fit is finished, only a second or two.
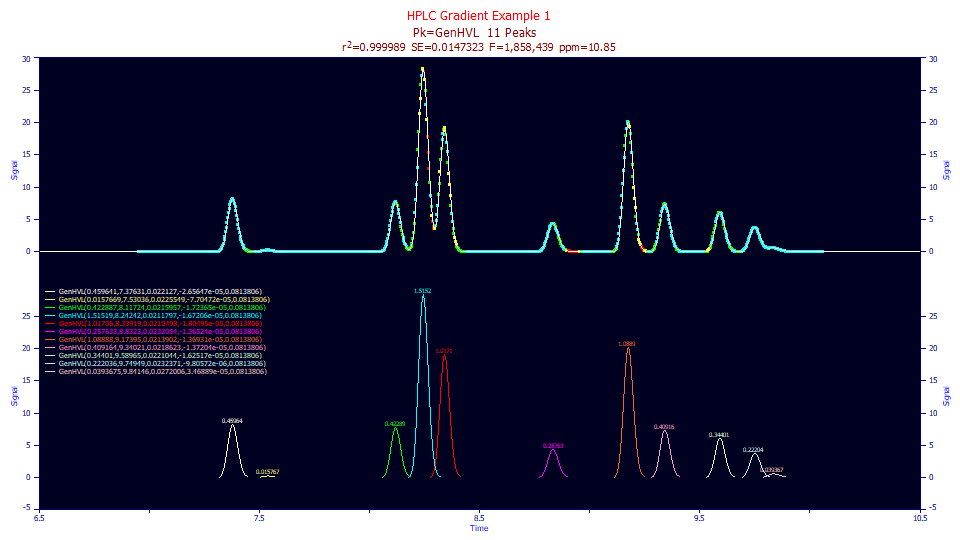
The 10.87 ppm error is good, but we will also fit the Gen2HVL model to see if there was too strong or too weak of an unwinding. The Gen2HVL model's a4 power of decay parameter should fit to very close to 2.0 if the unwinding was accurate.
Click Cancel to immediately return to the placement screen. Change the model to the Gen2HVL. Click Peak Fit, and then click OK. Click Review Fit when the fitting is finished.
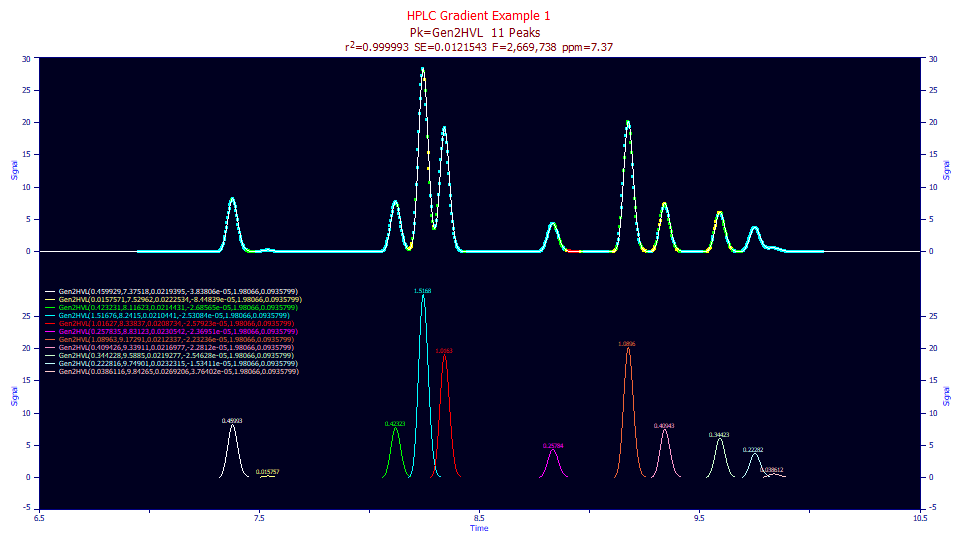
The 7.37 ppm error suggests we were fairly close to an optimal unwinding.
If needed, click on the Numeric button to open the numeric summary and under the Options menu, choose Select Only Fitted Parameters.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999263 0.99999224 0.01215434 2,669,738 7.37478257
Peak Type a0 a1 a2 a3 a4 a5
1 Gen2HVL 0.45992868 7.37517987 0.02193946 -3.838e-05 1.98065954 0.09357987
2 Gen2HVL 0.01575713 7.52961772 0.02225342 -8.448e-05 1.98065954 0.09357987
3 Gen2HVL 0.42323125 8.11622542 0.02144310 -2.686e-05 1.98065954 0.09357987
4 Gen2HVL 1.51676271 8.24150130 0.02104409 -2.531e-05 1.98065954 0.09357987
5 Gen2HVL 1.01627415 8.33837014 0.02087343 -2.579e-05 1.98065954 0.09357987
6 Gen2HVL 0.25783518 8.83123496 0.02305417 -2.37e-05 1.98065954 0.09357987
7 Gen2HVL 1.08963446 9.17290683 0.02123367 -2.232e-05 1.98065954 0.09357987
8 Gen2HVL 0.40942620 9.33910702 0.02169774 -2.281e-05 1.98065954 0.09357987
9 Gen2HVL 0.34422844 9.58850247 0.02192766 -2.546e-05 1.98065954 0.09357987
10 Gen2HVL 0.22281563 9.74900684 0.02323154 -1.534e-05 1.98065954 0.09357987
11 Gen2HVL 0.03861160 9.84264943 0.02692057 3.764e-05 1.98065954 0.09357987
The a4 power of decay in the Gen2HVL fit to a value of 1.98, very close to our expectation. Most isocratic peaks, in our experience, will fit to 1.97-1.99, just below below 2.0. This suggests the <*g*> convolution width of 0.0203 taken from the standards produces an adequate unwinding, neither too strong nor too weak.
Click Cancel to immediately return to the placement screen and OK to return to the main window.
Fitting the Gradient in Production Data
We assume your standards will properly give you the gradient estimate, but if such is not the case, and you need to extract the gradient unwind parameter(s) directly from messy production data, we can recommend three approaches for getting the gradient estimate directly from a production sample. We will finish this gradient peak set of tutorials with these three methods:
Isolate One or More Baseline Resolved Peaks
If you have baseline resolved peaks, you can use a feature of the baseline subtraction to remove all adjacent peaks and force the zero extended baseline the deconvolution modeling requires. This is the simplest of the three approaches, but a measure of intelligent preprocessing is needed.
Create a Series of Different Unwindings and Determine the Optimum from Goodness of Fit
In this approach, you must generate replicate data sets and in each create a different strength of unwinding. By fitting a model whose power of decay is 2, such as the GenHVL, the goodness of fit will be at a maximum when the unwinding is closest to complete, neither overdone or underdone. This is an approach that works irrespective of baseline resolved peaks. It can appear somewhat cumbersome, but it is effective and it is difficult to get an incorrect estimate, and with some experience, it actually requires very little time.
Fit a Deconvolution Model to the Production Data as Is, Painstakingly Jumping the Fourier-Introduced Local Minima
In this approach, you simply nurse the deconvolution model, applied to every peak in the production data, through the Fourier filter's oscillations in n-dimensional parameter space. No preprocessing is needed, only a good measure of patience and persistence.
Isolate One or More Baseline Resolved Peaks
In this approach we must identify baseline resolved peaks in the data and zero everything else, including all peaks which we wish to remove from the estimation process.
Selected Imported Data from the Data Levels dropdown in the main window. Click Copy, answer Yes, and OK to accept the default new data level titles. We now have a Copied Data(*) data level. The (*) means it will be replaced by a Transform, Baseline, or IRF Deconvolution level if you immediately engage one of these procedures.
Click Subtract Baseline.
We will be manually assigning baseline and zeroing those points, so the settings for automated baseline detection will not matter. We are still fitting extracted peaks which will be baseline corrected (even though this data has already been baseline corrected), so the baseline modeling will matter. Please be sure the Model dropdown is set to Non-Parm Linear with the default NP n of 10, the window size of the points used for the non-parametric fit. The requirement of consecutive points can be left at 10. It will not matter when manually setting the baseline as we will now do.
Check Zero Baseline Points and Zero Negative Points. Note that Zero Baseline Points and Zero Negative Points are not normally used options for conventional baseline processing.
![]() To help a bit with visualization, click the Zoom-In mode in the graph toolbar. Further zoom-in
the already zoomed baseline points as follows:
To help a bit with visualization, click the Zoom-In mode in the graph toolbar. Further zoom-in
the already zoomed baseline points as follows:
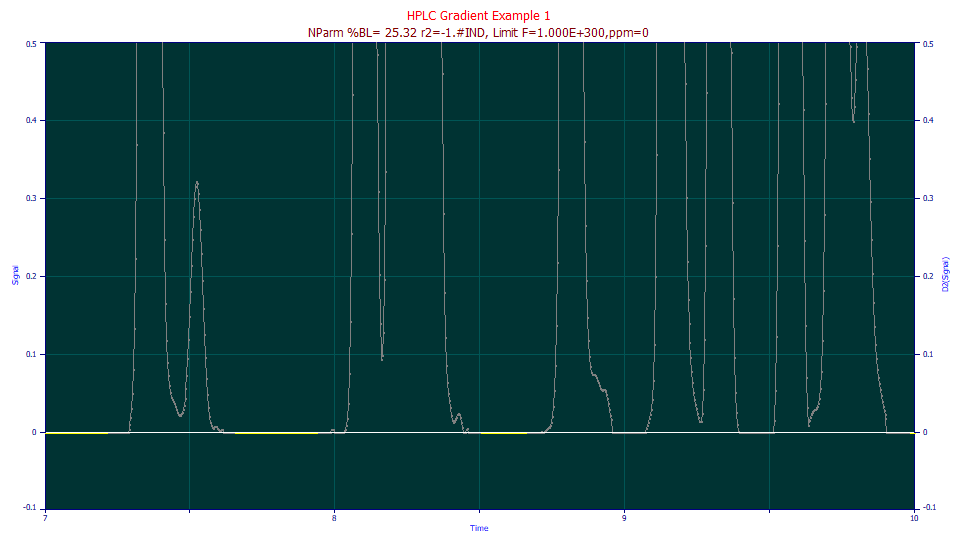
We agree, there isn't one peak you would even want to isolate. Still, we will employ this first approach for gradient estimation with what we have available.
![]() Click the Manual Sectioning mode. Be sure either the 2nd Deriv Zero or SD Variation
algorithm is selected. Manual baseline assignment is only available with these algorithms.
Click the Manual Sectioning mode. Be sure either the 2nd Deriv Zero or SD Variation
algorithm is selected. Manual baseline assignment is only available with these algorithms.
Baseline points are assigned (peak removed) by left clicking the mouse at a point where the baseline is deemed to conclude, and sliding left and releasing the mouse when all of the intended baseline points have turned yellow (instead of gray).
Peak points are assigned (baseline removed) by left clicking the mouse at a point where the peak information is deemed to begin, and sliding right and releasing the mouse when all of the intended peak points have turned gray (instead of yellow).
What we wish to do is make the three peaks at 7.52, 8.82, and 9.58 discrete and the only peaks in the data that emerge from this procedure.
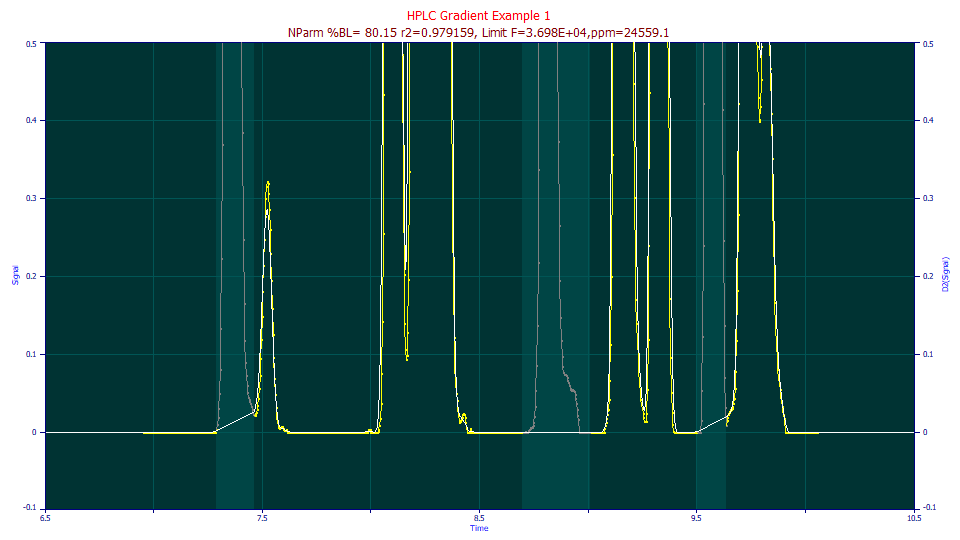
Reproduce as closely as you can the pattern of baseline and peak as above.
Start at the far right, click the mouse and slide to the far left, rendering the entirely of the data as baseline (all yellow). Place the mouse just before the start of the first peak that is grayed above, click and hold the mouse button down while sliding to the right and stop at the valley between the two peaks and release the mouse. For the second of the three peaks grayed above, start just before the peak begins to rise and stop just after the ugly feature in its tail ends. For the third peak grayed above, start just before the rise and again stop at the lowest point in the valley.
This does take some getting used to, as it is counterintuitive to assign major peaks as baseline. In actual practice, it only requires a few seconds.
If you look at the three gray peaks and the white baseline below them, you see that the first and last were not fully baseline-resolved, and the middle peak, which was, has a couple of very tiny components in its tail. We thus illustrate why other approaches to estimating the gradient with production data may have value. Even though the premise of isolating one more baseline-resolved peaks failed, we will proceed with the analysis.
One you have the assignments similar to the above, click OK, and click OK to acknowledge the manual processing.
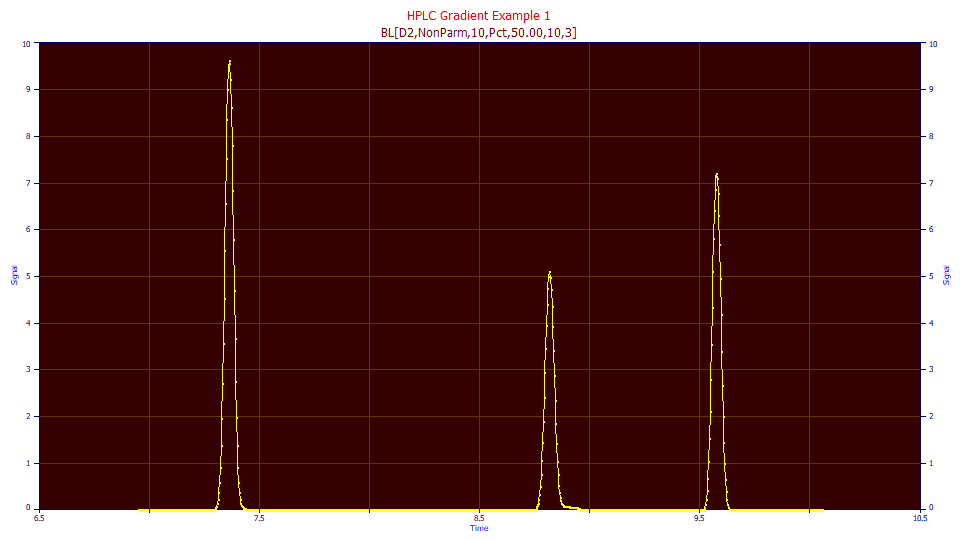
Exactly as we did in the second tutorial, click the Local Maxima Peaks button.
Be sure the following settings are in place. We will fit the GenHVL<*g*> deconvolution model to get the gradient's half-Gaussian SD for these four peaks.
Peak Detection
Set Sm
n(1) to 5
Peak Type
Select Chromatography
w/IRF in the first dropdown
Select GenHVL<*g*> as the model in the
second dropdown
Scan
Set the Amp %
threshold to 1.5 %
Leave Use Baseline Segments unchecked
Be sure Use IRF,ZDD is checked
Vary
Leave width a2
and shape a3 checked, all other unchecked
If the IRF value for the <g> SD is the .02 from the prior tutorial or the default, the Gaussian IRF deconvolution width will be too high. There will again be these oscillations below zero from the deconvolution.
Click the IRF button and for the <g> IRF, enter 0.01 for the g width (sd) default starting estimate. Click OK.
Click Peak Fit to open the fit strategy dialog. Select the Fit with Reduced Data Prefit, Cycle Peaks, 2 Pass, Lock Shared Parameters on Pass 1. Leave the Fit using Sequential Constraints box checked. Click OK.
Click Review Fit.
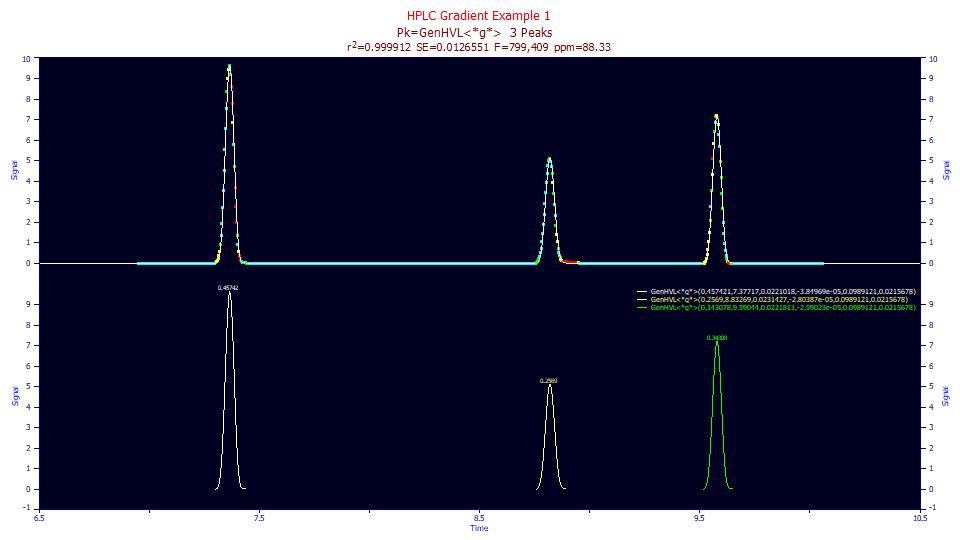
The fit is weak, but such was not unexpected.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99991167 0.99991032 0.01265508 799,409 88.3265916
Peak Type a0 a1 a2 a3 a4 a5
1 GenHVL<*g*> 0.45742137 7.37717205 0.02210176 -3.85e-05 0.09891214 0.02156776
2 GenHVL<*g*> 0.25689977 8.83269307 0.02314267 -2.804e-05 0.09891214 0.02156776
3 GenHVL<*g*> 0.34307824 9.59044454 0.02218133 -2.59e-05 0.09891214 0.02156776
The three peaks have a shared a5 half-Gaussian deconvolution SD of 0.0216, in reasonable agreement with the 0.0203 average from the fits of the standards.
Click OK to close the Review, OK to accept a saved GenHVL<*g*> fit in the data file, OK to acknowledge the revision in the file, OK to return to the placement screen, and OK to return to the main window.
Creating an Optimization
The following steps are a useful exercise if you find you need to create an optimization experiment based on fitting replicated data sets with differences in preprocessing.
We will replicate the baseline-corrected data and generate different <g> convolution unwound data sets. If these sets are fit to models with only a skewness third moment adjustment, such as the GenHVL, where there is no fourth moment power of decay adjustment impacting the thinness or fatness of the tailing, this intrinsic power of 2.0 decay will cause the goodness to fit to an optimum where the gradient unwinding most closely accommodates the model. We want to fit a model whose goodness of fit will be adversely impacted by the lack of an optimal unwinding of the gradient, not a model that capably adjusts for unwinding that is either too weak or too strong.
With Imported Data selected at the main window, select New to create a new PeakLab data file with only this data level. Yes to proceed. Yes to save the prior data if asked to do so. Note that the newly created file is not yet saved.
Right click the graph and select Replicate this Data Set a Specified Number of Times from the popup menu. Enter 10 to create 10 replicates. There are now 11 identical data sets.
Click the IRF Deconvolution procedure button and be sure the following settings are in place:
Response Fn
<g> Gaussian
Convolve Right (we want convolution, not deconvolution)
IRF Parameters
0.0203 (the
value from the GenHVL<*g*> deconvolution integral fit
Genetic Alg Optimization
These settings will not be used
Fourier Filter
None (no filtering is generally needed for convolution)
Post-Processing
Zero Baseline Points - unchecked (baseline will be
actual subtracted values)
Zero Negative Points - unchecked (negative
baseline points are permitted)
Double click the first data set and enter a custom unwind <g> convolution width of 0.025. Double click to return to all data sets with this set retaining the custom adjustment. The first data set will show a <g> [0.025] in its graph title. Repeat this custom adjustment for each of the data sets. Double click the second data set, enter 0.024, and double click again to restore all of the graphs. Do the same for the third data set, enter 0.023, and for the fourth 0.022, and so on until the last of the 11 data sets is given the custom unwind width of 0.015.
Click OK, OK to acknowledge the local convolution warnings, and OK to accept the titles. There is now a 'DC' data level with 11 different <g> unwind convolution widths. And once more the tedious work is finished, and the program will do the rest.
Click the Hidden Peaks - Second Derivative button. We still need the hidden peak method for the higher deconvolution widths in the series which wash out the local maximum in the last tiny peak rendering it hidden. Leave everything as set in the prior fitting except change the model to the GenHVL. Again we need a model that enforces a power of decay of 2.0 in order for this optimization to work. You will see 11 peaks automatically placed in each of the 11 data sets.
Click Peak Fit to open the fit strategy dialog. Be sure the Fit with Reduced Data Prefit, Cycle Peaks, 2 Pass, Lock Shared Parameters on Pass 1 and Fit using Sequential Constraints boxes are checked. Click OK. Click Review Fit when the fitting is finished.
Right click any one of the graphs in the Review and select the Map Experimental Process Variables for Fitted Data Sets option. Enter <g> for the variable name and the values 0.025, 0.024, etc. for the 11 sets. Click OK.
Click Explore, set <g> as the x variable and Fit Stats for the y variable.
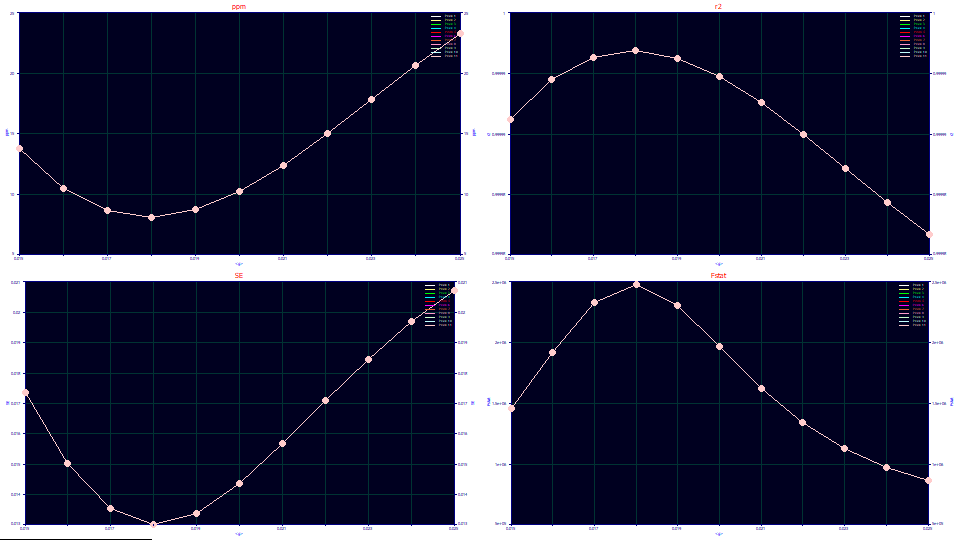
For this production sample, the <g> optimal unwind width for a GenHVL fit is less, about 0.018 instead of the 0.020 we saw with the fitted standards. At this optimum, the ppm is at a minimum (the r2 at a maximum), the SE is at a minimum, and the F-statistic at a maximum. It is straightforward to estimate 0.0135 as very close to the optimum <g> unwind width.
This optimization is accurate because we optimized the very process we will use for the unwound fitting. In fact, if we are willing to use the 0.018 value for the unwinding, we are already finished:
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999187 0.99999145 0.01301282 2,476,110 8.13218881
Peak Type a0 a1 a2 a3 a4
1 GenHVL 0.45978603 7.37358446 0.02135035 -3.489e-05 0.07836035
2 GenHVL 0.01572565 7.52809144 0.02171820 -7.995e-05 0.07836035
3 GenHVL 0.42301167 8.11444321 0.02079366 -2.54e-05 0.07836035
4 GenHVL 1.51605097 8.23966666 0.02037827 -2.438e-05 0.07836035
5 GenHVL 1.01675991 8.33650315 0.02022356 -2.507e-05 0.07836035
6 GenHVL 0.25757480 8.82958350 0.02243447 -2.093e-05 0.07836035
7 GenHVL 1.08917613 9.17115589 0.02058068 -2.092e-05 0.07836035
8 GenHVL 0.40939675 9.33760412 0.02109691 -1.963e-05 0.07836035
9 GenHVL 0.34424315 9.58704329 0.02135228 -2.194e-05 0.07836035
10 GenHVL 0.22224651 9.74721598 0.02254189 -1.36e-05 0.07836035
11 GenHVL 0.03908607 9.83991249 0.02638390 3.4451e-05 0.07836035
Here we do see the the unwound peaks fitted to an isocratic model with a higher degree of accuracy than fitting the compressed shapes. Is there a difference between 8 ppm and 20 ppm unaccounted variance? The better question pertains to the difference between fitting the compressed shapes with a fourth moment isocratic model, and unwinding the gradient to produce an isocratic peak which is then fitted to model with only a third moment skewness adjustment.
As we noted prior, the unwound peaks will have a higher a2 width.
Unwinding the peaks to a deconvolved isocratic shape does mean the a3 chromatographic shapes should be more accurate. In the first gradient tutorial, we saw almost no correlation between the a3 and the different moments in these direct fits of the compressed gradient shapes. In the second gradient tutorial, a3 had a greater impact on moments when the gradient was modeled and some form of isocratic unwinding occurred. If we omit the two tiny peaks, the a3 values vary here from -3.49e-5 to -1.36e-5, all fronted, and all in a very narrow range. The Gen2HVL fits to the gradient shapes had a mix of +a3 and -a3 values varying from -8.7e-5 to +9.2e-5. (fronted to tailed), and we know from the fits to the standards that all should be fronted (-a3).
Click OK to close the Review, OK to accept a saved GenHVL fit in the data file, OK to acknowledge the revision in the file, OK to return to the placement screen, and OK to return to the main window.
Close this data file with the replicates.
Creating the new Data Layer with the Optimized Unwinding in Original File
We will create a new data level with the <g> convolution unwind width of .018.
Select the file HPLCGradientPeaks.pfd from the bottom of the MRU (most-recently used) list of files in the File menu.
Select the Imported Data in the Data Levels dropdown and click the Copy button. Answer Yes to create the new data level and OK to accept the default titles. Click the IRF Deconvolution button and change the <g> parameter value to 0.0135. Click OK to accept the convolution, and OK once again to accept the data level titles.
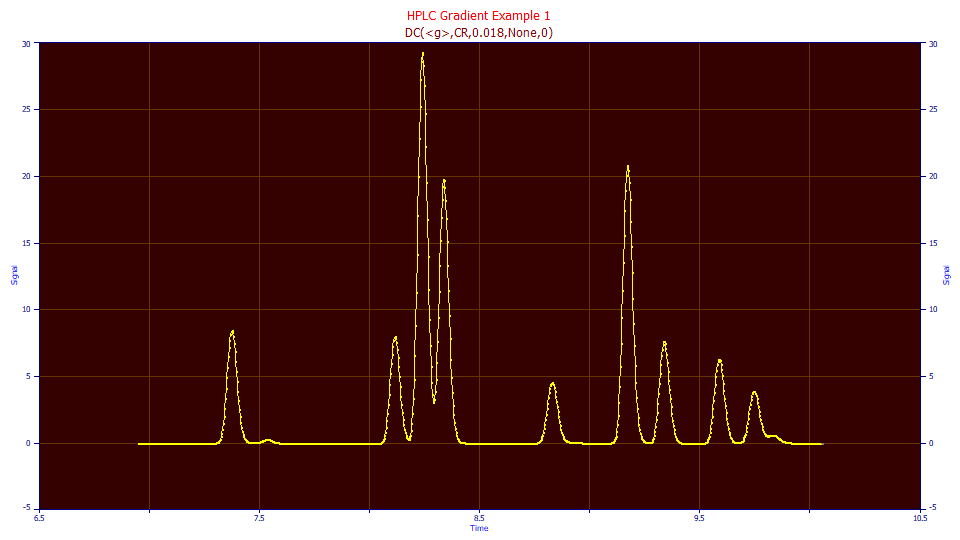
Since the peaks retained their local maxima with this 0.018 <g> response function convolution width, though barely, we no longer need to use a hidden peak method.
Click the Local Maxima Peaks button. From the Chromatography family, change the model to the GenHVL. Set the Amp % threshold to 0.5 %. In the Vary section, be sure the a2 and a3 are checked and that a4 is unchecked. You should see the 11 peaks, all automatically placed.
Click Peak Fit to open the fit strategy dialog. With the Fit with Reduced Data Prefit, Cycle Peaks, 2 Pass, Lock Shared Parameters on Pass 1 and Fit using Sequential Constraints boxes checked, click OK.
Click Review Fit when the fit is complete.
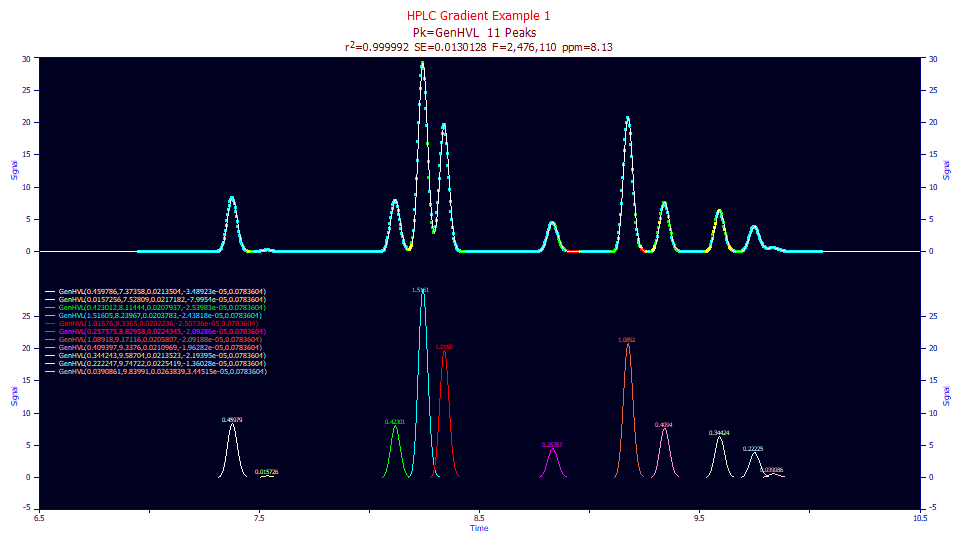
The error with the 0.18 fit is 8.13 ppm, and we again contrast this with the 19.96 ppm on the Gen2HVL direct fits of the compressed shapes.
If needed, Click on the Numeric button to open the numeric summary and under the Options menu, choose Select Only Fitted Parameters and then check Chromatographic Analysis.
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999187 0.99999145 0.01301282 2,476,110 8.13218881
Peak Type a0 a1 a2 a3 a4
1 GenHVL 0.45978603 7.37358446 0.02135035 -3.489e-05 0.07836035
2 GenHVL 0.01572565 7.52809143 0.02171820 -7.995e-05 0.07836035
3 GenHVL 0.42301167 8.11444321 0.02079366 -2.54e-05 0.07836035
4 GenHVL 1.51605097 8.23966666 0.02037827 -2.438e-05 0.07836035
5 GenHVL 1.01675991 8.33650315 0.02022356 -2.507e-05 0.07836035
6 GenHVL 0.25757480 8.82958350 0.02243447 -2.093e-05 0.07836035
7 GenHVL 1.08917613 9.17115589 0.02058068 -2.092e-05 0.07836035
8 GenHVL 0.40939675 9.33760412 0.02109691 -1.963e-05 0.07836035
9 GenHVL 0.34424315 9.58704329 0.02135228 -2.194e-05 0.07836035
10 GenHVL 0.22224651 9.74721598 0.02254189 -1.36e-05 0.07836035
11 GenHVL 0.03908607 9.83991249 0.02638390 3.4451e-05 0.07836035
Click OK to close the Review, OK to accept a saved GenHVL fit in the data file, and OK to acknowledge the revision in the file. Click OK to once again return to the main window.
Patiently Fitting a Deconvolution Model to the Production Data
Select Imported Data from the Data Levels dropdown.
Click Local Maxima Peaks.
We will now fit the GenHVL<*g*> deconvolution directly to this data. This is a useful exercise insofar as you will see firsthand the very significant differences between discrete convolution and deconvolution.
![]() Click Modify Peak Fit Preferences. Change the maximum total iterations to 2000. You will see soon
enough why we increased the iterations. Click OK.
Click Modify Peak Fit Preferences. Change the maximum total iterations to 2000. You will see soon
enough why we increased the iterations. Click OK.
Select the Chromatography with IRF peak family and the GenHVL<*g*> deconvolution model. Click IRF, and set the <g> IRF g width (SD) parameter value to 0.025. Click OK.
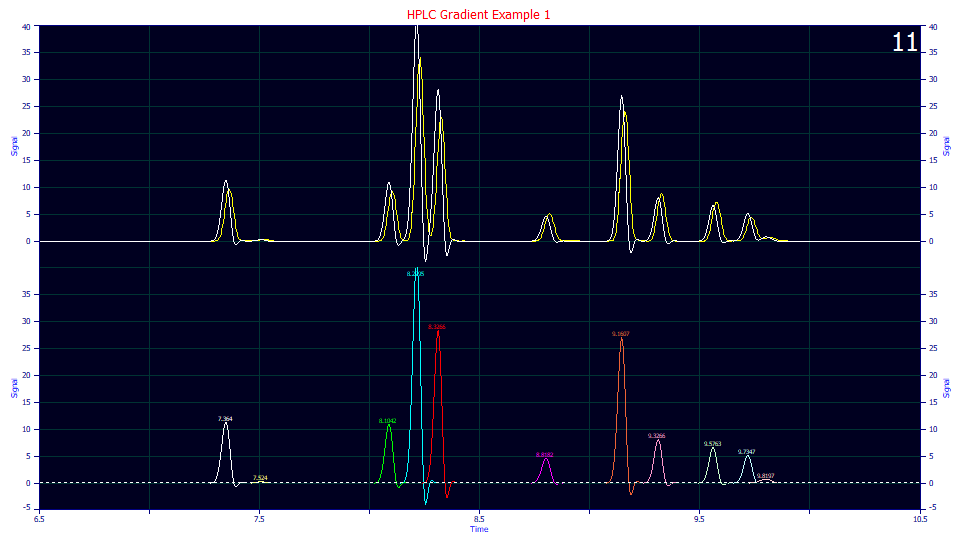
Look closely at the peaks in the lower plot. If we were to lock this 0.025 value we would have deconvolutions with this negative oscillation that no amount of adjustment in the other parameters can correct. An adjacent peak can sometimes increase in amplitude in an attempt to offset this negative oscillation, but the fitted values will lose validity.
Click IRF, and set the <g> IRF's g width (SD) parameter value to 0.018 and uncheck its Lock button, if needed. We will (attempt to) fit the <g> width. Click OK. The negative oscillations are no longer visually apparent. Keep these oscillations in the baseline of the deconvolved peaks in mind as we proceed.
Click Peak Fit to open the fit strategy dialog. With the Fit with Reduced Data Prefit, Cycle Peaks, 2 Pass, Lock Shared Parameters on Pass 1 and Fit using Sequential Constraints boxes checked, click OK.
The fit will battle local minimum in the 46-dimensional parameter space and finish at about 105 ppm. Click Review Fit. Click Numeric if needed, and note that the a5 half-Gaussian fit to 0.01788. Click OK. Choose Cancel in the Save. Restart the fitting.
We will run another set of 2000 iterations with the algorithm fighting local minima arising from oscillations in the Fourier noise filter for each of the peaks. The fit should stop at around 75 ppm. Once again, click Review Fit. Click Numeric if needed, and note that the a5 half-Gaussian fit is now 0.01785. Click OK. Choose Cancel in the Save. Restart the fitting.
When peaks lack a clear baseline to each side, the Fourier deconvolution and filtering will produce oscillations at the baseline which produce local minima that the PeakLab algorithm must seek to resolve. . The next stop is around 52 ppm. Once again, click Review Fit. Click Numeric if needed, and note that the a5 half-Gaussian fit is now 0.01788. Although the algorithm has not converged, the estimated parameter for the gradient has mostly done so.
In the Review, zoom-in the baseline of the lower plot.
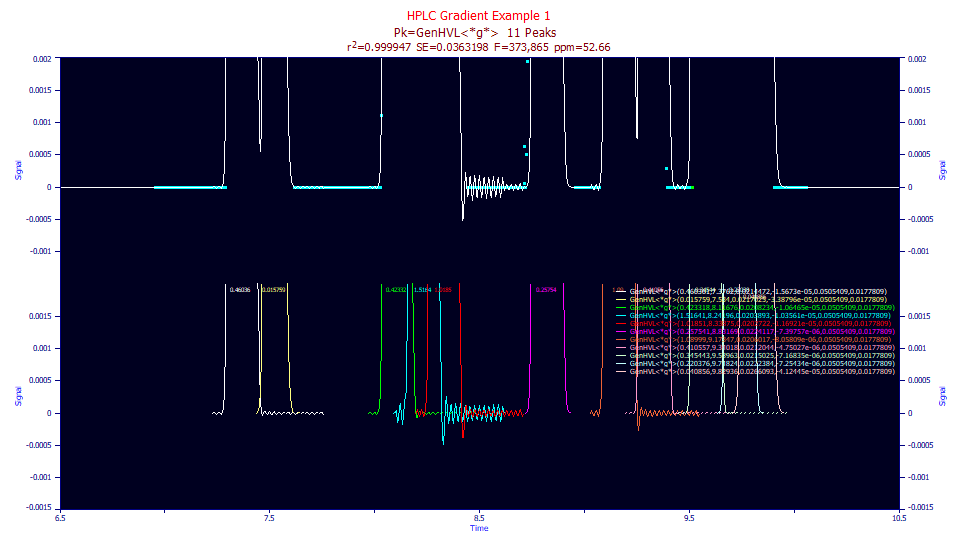
Note that most of the peaks have Fourier oscillations in the baseline. This is why we preprocessed the data as we did in the first approach, even though we could not find even one clean baseline-resolved peak. We sought to estimate the gradient free of these oscillations.
Click OK to close the Review, OK to accept a saved GenHVL<*g*> fit in the data file, OK to acknowledge the revision in the file, OK to return to the placement screen, and OK once again return to the main window.
Convolution Genetic Optimization
There may also be any number of other ways to estimate the gradient in a production sample with many peaks.
We will also share an experimental estimation method, and we invite you to share with us any that you may discover.
If we add the Deconvolved Moments to the Numeric Summary in the second gradient tutorial 's fits of the standards, and we look at the averages of the 8 fits we will see the following:
Average for 8 Fits
Fitted Parameters
r2 Coef Det DF Adj r2 Fit Std Err F-value ppm uVar
0.99999462 0.99999427 0.07764688 11,487,966 5.37634646
Peak Type a0 a1 a2 a3 a4 a5
1 GenHVL<*g*> 6.89666917 8.62628210 0.02141637 -2.809e-5 0.09235604 0.02032334
Measured Values
Peak Type Amplitude Center FWHM Asym50 FW Base Asym10
1 GenHVL<*g*> 152.467810 8.61343184 0.04407221 0.97967742 0.08458621 0.98022360
Deconvolved Moments
Peak Type Area Mean StdDev Skewness Kurtosis Amplitude Center
1 GenHVL 6.89641373 8.62945961 0.00047681 0.20134307 3.06903946 129.615481 8.62773776
The key values we need are the Amplitude of the convolution (the GenHVL) and of the eluted peak (the GenHVL<*g*>). The average amplitude of the compressed gradient peaks is 152.4, the average amplitude of the fitted unwound peaks is 129.6. For these standards peaks, the average amplitude attenuation is 0.85. If you have an expectation for amplitude attenuation, you can use this in a genetic algorithm convolution estimation of the gradient.
Select the Imported Data in the Data Levels dropdown and click the Copy button. Answer Yes to create the new data level and OK to accept the default titles.
Click the IRF Deconvolution button.
Response Fn
<g> Gaussian
Convolve Right (we want convolution, not deconvolution)
IRF Parameters
0.0203 (the
value from the GenHVL<*g*> deconvolution integral fit
Genetic Alg Optimization
Parameter 1 % 90
(parameter will vary 0.00203-0.03857)
AmpAtten, 0.85
Fourier Filter
None (no filtering is generally needed for convolution)
Post-Processing
Zero Baseline Points - unchecked (baseline will be
actual subtracted values)
Zero Negative Points - unchecked (negative
baseline points are permitted)
For convolution optimizations, the genetic algorithm seeks to find the same amplitude attenuation and also maximize the amount of baseline. In this case, with just one parameter, it is mostly the specification of an amplitude attenuation. We do allow a certain band within the attenuation where we search for maximized baseline.
Click the Genetic Optimize button.
You should see something like <g> [0.187] in the second graph title. By putting in the amplitude attenuation of 0.85 which we saw on the 0.02 <g> standards peaks, and optimizing with this generic algorithm on the production data, we see a <g> of 0.0187, reasonably close to our 0.018 optimization and the 0.0230 from the standards averaging.
Let's see what AmpAtten is needed in this genetic procedure to replicate the 0.0203 <g> width we saw with the standards fits, those which produced the average 0.85 amplitude attenuation in their GenHVL<*g*> fits. Enter .832 in the AmpAtten field again click Genetic Optimize.
You now see <g> [0.0203] in the second graph title.
For any estimation procedure to be viable, the fits to the unwound data must be superior in goodness of fit to the Gen2HVL fits of the compressed data as eluted.
Considerations
Creating Replicate Data Sets Each Preprocessed with Different Unwind Values
The replication of the data and the application of different <g> convolution widths to such data sets, followed by GenHVL fits with a goodness of fit optimization, should work for any data set. The convolutions decrease noise and the GenHVL model is exceptionally stable. You may need to use a hidden peak method or manually place the peaks that cease to be local maxima peaks after the convolution.
Fitting the Gradient with a Deconvolution Model
The GenHVL<*g*> fitting of the gradient will be very dependent on the extent to which the peaks are baseline resolved. In our production data example, there wasn't one clean isolatable peak with a good measure of baseline to each side that we could extract as if it were a standard. If you must fit all of the data containing many peaks, the Fourier baseline oscillations will likely play havoc with convergence to the global minimum. You may have to restart the fits a number of times to escape a host of local minima, which will be introduced by the Fourier deconvolution and noise filtration.
Compare the Gen2HVL Fit of the Compressed Data
As a crosscheck on the unwinding value and constancy of the gradient across the peaks, we recommend also fitting the Gen2HVL to the compressed gradient shapes. If the unwound fit has a weaker goodness of fit, check to see if the unwinding caused a small local maximum peak to become a hidden peak which was not accounted in fitting the unwound data. The reason data is unwound is to realize more accurate estimates of the true parameters of the peak absent the presence of the gradient. That improved accuracy depends not only on a correct unwinding of the gradient, but also on managing each peak as successfully in this unwound shape as in its compressed state. Also, by fitting this Gen2HVL to the eluted shapes, you will get a separate peak by peak estimate of the gradient strength as it impacts the fourth moment tailing in each compressed peak. When you unwind a gradient for fitting, you are globally unwinding the whole data set with a single specification for the gradient.
When you fit the Gen2HVL to the eluted data, and a model such as the GenHVL to the unwound data, you have two sets of fits which tell you everything you can model about the chromatography producing the peaks. If you have a data level with the compressed peaks, and one with the unwound data, you can readily refit either as needed, or create a new data level with a different unwinding. For gradient data, we recommend that you always retain the compressed shapes in an original Imported Data level and that you have a Gen2HVL fit of that data on a subsequent data level where the baseline has been subtracted.


 |